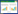文章信息
- 刘元望, 李兆君, 冯瑶, 成登苗, 胡海燕, 张文娟
- LIU Yuan-wang, LI Zhao-jun, FENG Yao, CHENG Deng-miao, HU Hai-yan, ZHANG Wen-juan
- 微生物降解抗生素的研究进展
- Research progress in microbial degradation of antibiotics
- 农业环境科学学报, 2016, 35(2): 212-224
- Journal of Agro-Environment Science, 2016, 35(2): 212-224
- http://dx.doi.org/10.11654/jaes.2016.02.002
-
文章历史
- 收稿日期: 2015-06-15
2. 山西师范大学地理科学学院, 山西 临汾 041004
2. College of Urban and Environment Science, Shanxi Normal University, Linfen 041004, China
抗生素(Antibiotics)是由微生物(包括细菌、真菌、放线菌)产生的具有抗病原体或其他活性的一类次级代谢产物,能干扰其他活细胞的发育功能。抗生素作为抑菌或杀菌类药物已被广泛应用于人类疾病治疗、畜禽及水产养殖等多个领域,主要包括四环素类、磺胺类、β-内酰胺类、氟喹诺酮类和大环内酯类等[1]。我国是抗生素生产和使用大国,每年抗生素生产量达21万t,使用量达18.9万t,其中兽用抗生素占到使用量的一半以上[2]。研究发现,生物体摄入大量抗生素类药物后除部分被机体代谢外,有40%~90%以原药或初级代谢产物的形式随粪便和尿液排出体外[3],最终又通过施肥等方式进入土壤环境或者通过渗漏和污水排放进入水体环境。近年来,关于抗生素类污染物在水体、沉积物和土壤中被检出的国内外相关报道层出不穷,甚至在蔬菜、奶类和肉类等产品中也发现了抗生素残留[4, 5, 6, 7, 8]。
研究发现,长期暴露在抗生素环境下,不仅人和动物的患病和发病率会升高,而且对植物的叶绿素合成、酶分泌和根系生长都有影响[9, 10]。此外微生物也会逐渐适应抗生素环境,并产生抗生素耐药性和抗性基因(Antibiotic resistance genes,ARGs)[11, 12]。同时,低浓度抗生素对生态环境中微生物种群也能够起到筛选作用,使具有抗生素耐药性的微生物种群得以保留并逐渐壮大,而对其敏感的种群不断死亡消失,直接后果就是使微生物种群结构失衡,对生态环境及人类健康造成极大的危害。据报道,2014年全球有70万人因抗生素耐药性的产生而死亡,因此解决抗生素问题迫在眉睫[13]。
为了解决抗生素污染问题,除了减少抗生素的滥用,如何去除环境体系中残留的抗生素已经成为近年来研究的热点。目前,对含有抗生素残留污水的理化处理方法已进行了大量的研究和实践[14, 15],包括高级氧化法、活性炭吸附法、低温等离子体技术和膜处理法等[16]。但是这些理化法处理所需成本高、管理复杂,除了高级氧化法对抗生素的去除率可达95%外,其他方法的去除效率都较低,并且都对固态介质中抗生素残留处理存在局限性。因此,有关抗生素的微生物降解研究逐渐成为热点[17]。本文基于近年来抗生素污染微生物处理方法、抗生素降解功能菌的筛选、降解条件、降解效果和降解机制方面的研究进行了系统的综述,旨在为后续抗生素微生物降解研究提供参考。
1 微生物处理方法 1.1 活性污泥法活性污泥法(Activated sludge process,ASP)是国内外处理抗生素污水最常见的方法。利用活性污泥消除污水中抗生素的方法一般包括物理吸附(腐殖质、活性炭、絮凝剂)、化学反应和微生物降解。ASP发展时间早,工艺成熟,积累了大量的运行和管理经验。因此该方法经常用于含抗生素废水的处理。
四环素类抗生素(TCs)在活性污泥中的去除主要以吸附为主,微生物降解较小甚至不存在微生物降解[18, 19, 20, 21]。与TCs不同,磺胺类抗生素(SAs)在ASP中的去除主要是微生物降解起作用[22, 23, 24],但是不同的SAs降解效果不尽相同。Yang等[25]研究发现,在相同降解条件下,磺胺间甲氧嘧啶(SMM)降解率为19%,磺胺甲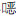 唑(SMX)为24%,而磺胺二甲嘧啶(SDM)为30%。污泥龄(SRT)和反应时间会显著影响SAs的降解效果[25]。不同SRT和反应时间对磺胺甲嘧啶(SMZ)降解效果影响的研究结果显示,随着SRT由5 d延长到25 d,SMZ的去除率可以由45%提高到80%;SMZ在活性污泥处理0.5~4.5 h内降解效果有显著差异[23]。Yang等[25]还发现在ASP中SAs的降解呈S型曲线,前期(2 d或3 d内)SAs降解缓慢,直到12 d降解比较稳定,降解率可达到95%,14 d后降解基本完成。这可能是由于微生物的适应过程,也可能是由于存在其他容易降解的异型生物质与SAs的降解发生竞争作用所致。温度也是影响SAs降解的主要因素。研究发现在20 ℃时SAs的降解迟滞期短,降解率高;而6 ℃时迟滞期会延长4倍左右,降解率低[24]。除此之外,由于SAs可以作为活性污泥中微生物的碳源或者氮源,活性污泥中碳源和氮源的含量会影响其降解效果。Müller等[27]通过设置不同的共代谢基质,发现在ASP系统中,增加碳源和减少氮源均可以提高SMX的降解效果。通过以下方式可以提高ASP对SAs的降解效率:(1)针对不同的SAs筛选不同的高效降解菌;(2)通过加入胞外聚合物提高微生物对抗生素的获得能力来促进抗生素降解[25];(3)优化并选择适合ASP中微生物团体生长和SAs降解的温度;(4)在加入到ASP系统前,将活性污泥中的微生物团体在相似的环境下进行适应性生长训练;(5)控制SRT,当SRT达到SAs的降解瓶颈时更新污泥;(6)针对不同的SAs和微生物团体优化ASP系统中的营养基质。
唑(SMX)为24%,而磺胺二甲嘧啶(SDM)为30%。污泥龄(SRT)和反应时间会显著影响SAs的降解效果[25]。不同SRT和反应时间对磺胺甲嘧啶(SMZ)降解效果影响的研究结果显示,随着SRT由5 d延长到25 d,SMZ的去除率可以由45%提高到80%;SMZ在活性污泥处理0.5~4.5 h内降解效果有显著差异[23]。Yang等[25]还发现在ASP中SAs的降解呈S型曲线,前期(2 d或3 d内)SAs降解缓慢,直到12 d降解比较稳定,降解率可达到95%,14 d后降解基本完成。这可能是由于微生物的适应过程,也可能是由于存在其他容易降解的异型生物质与SAs的降解发生竞争作用所致。温度也是影响SAs降解的主要因素。研究发现在20 ℃时SAs的降解迟滞期短,降解率高;而6 ℃时迟滞期会延长4倍左右,降解率低[24]。除此之外,由于SAs可以作为活性污泥中微生物的碳源或者氮源,活性污泥中碳源和氮源的含量会影响其降解效果。Müller等[27]通过设置不同的共代谢基质,发现在ASP系统中,增加碳源和减少氮源均可以提高SMX的降解效果。通过以下方式可以提高ASP对SAs的降解效率:(1)针对不同的SAs筛选不同的高效降解菌;(2)通过加入胞外聚合物提高微生物对抗生素的获得能力来促进抗生素降解[25];(3)优化并选择适合ASP中微生物团体生长和SAs降解的温度;(4)在加入到ASP系统前,将活性污泥中的微生物团体在相似的环境下进行适应性生长训练;(5)控制SRT,当SRT达到SAs的降解瓶颈时更新污泥;(6)针对不同的SAs和微生物团体优化ASP系统中的营养基质。
氟喹诺酮类抗生素在ASP中的微生物降解是其去除的次要途径,氧化还原条件、抗生素种类和污泥的含盐量等都会影响其降解效果。研究发现,在厌氧条件下氟喹诺酮的降解微不足道,在好养条件下降解率为14.9%~43.8%,在硝化条件下降解率为36.2%~60.0%,加入硝化抑制剂会显著减少氟喹诺酮的降解[29],并且淡水中氟喹诺酮不存在微生物降解,而在含盐污水中降解率可达到40.8%[21]。所以可以通过筛选高效降解菌株、提高通气量、加入硝化试剂或提高含盐量的方法来提高氟喹诺酮类抗生素在ASP中的降解率。
β-内酰胺类抗生素在ASP中的降解不够完全,尤其是在高浓度条件下降解率更低。Guo等[30]比较了Fenton、ASP和Fenton-ASP对阿莫西林的降解效果。研究结果显示,高浓度条件下单独采用ASP处理阿莫西林去除效果较差,而采用Fenton氧化去除率可达80%。将二者联合起来,即先用Fenton法处理,再用ASP处理,则最终可将阿莫西林完全降解[25]。
ASP尤其是ASP好氧处理法存在动力消耗大、处理成本高和易出现污泥膨胀现象等缺点[16, 31],其应用受到一定的限制。
1.2 膜生物反应器法膜生物反应器(Membrane bioreactor,MBR)是一种将薄膜对污染物的高效分离与微生物对污染物降解能力相结合的新型污水处理系统。这种方法采用超滤膜组件代替传统活性污泥工艺中的二沉池,可以进行高效的固液分离,克服了传统活性污泥工艺中出水水质不稳定、污泥容易膨胀等问题。此外,MBR还具有工艺参数容易控制、设备容积负荷高、占地少、性能稳定、易于自动控制管理等优点[32, 33]。较传统活性污泥工艺而言,MBR明显提高了污水中抗生素的去除效果。Sahar等[34]研究表明,MBR比传统活性污泥对大环内酯类抗生素、SAs和甲氧苄氨嘧啶类抗生素的去除率提高了15%~42%;Shen等[35]研究表明MBR对氨苄青霉素去除率比活性污泥法去除率高23%。可能是由于生物薄膜提高了对抗生素和生物量的保留作用,增加了微生物与抗生素的接触时间。
影响MBR对抗生素降解效果的因素主要包括抗生素种类、抗生素浓度、固体悬浮物含量(MLSS)、温度、化学需氧量(COD)、水力停留时间(HRT)和SRT等[36]。在MBR中即使是同一类别不同种类的抗生素去除效果也存在较大差异,有的去除率可达100%,有的去除率甚至为零[37]。这可能是由于流入MBR的污水中含有抗生素代谢物的离子,这些离子最终又会合成该种抗生素母体的缘故。当浓度不同时,抗生素的降解率也有所不同。研究发现,当浓度为50 ng·mL-1时,SAs在5 d降解率就达到90%以上,而浓度为1000 ng·mL-1时SAs的降解率很低。但是,不同浓度处理的SAs降解量相近,表明参加抗生素降解的酶具有类特异性[37]。一般较高含量的MLSS、较高的温度和较低的初始COD值均有利于抗生素的降解[38]。HRT和SRT会影响MBR对抗生素的降解,一般随着HRT和SRT的增加,抗生素的降解率会有相应的提高[32]。另有报道,β-变型杆菌和 γ-变形菌是污水处理过程中对抗生素去除起主要作用的菌,且随着SRT的增加,抗生素抗性基因呈现增加趋势,并且抗生素去除率有所提高。这可能是由于较长的HRT和SRT能够为微生物(如硝化菌和抗生素降解菌等)提供更多富集时间和空间的缘故[39, 40, 41]。
为了提高MBR工艺的降解效率,可以从以下几方面改进:(1)针对不同的抗生素筛选出具有高效降解能力的菌株;(2)在一定范围内提高MLSS的含量;(3)在一定范围内提高处理温度;(4)对要处理的废水首先进行降低COD的前处理;(5)相对增加HRT和SRT;(6)将MBR和其他方法联用[42, 43];(7)优化滤膜性能,根据不同净水要求选择不同类型膜组件。
1.3 超声生物法超声法是近几年来发展起来的一种新型的污水处理方法,正日益受到人们的关注。该法主要是通过超声波使液体中的微小泡核激化产生高温和高压,破坏抗生素的分子结构,从而达到降解目的。并且水分子在高温高压下产生诸如 H2O2和·OH等活性氧物质(Reactive oxygen species,ROS),氧化抗生素从而达到降解抗生素的目的。这可能是由于 H2O2和·OH等的链式反应能够氧化抗生素所致,因此在污水中加入诸如 Fenton试剂、H2O2、CH3Cl、臭氧等可以产生ROS的助剂以促进反应的进行[44, 45, 46, 47, 48, 49, 50, 51]。但是Lastre-Acosta等[46]却证明H2O2会抑制超声法对磺胺嘧啶的降解作用,这可能与抗生素的种类有一定关系。此外,在一定范围内超声功率越大、溶液 pH值越高(6~11)、气水比越大、抗生素浓度越低则超声法对抗生素的降解率越高[48, 49, 50, 51, 52]。但是也有研究表明,在中性或酸性条件下,超声法也能够获得较高的抗生素降解速率[47]。例如,Wei等[50]通过试验证明,在 pH为 7.2时利用超声法对左氧氟沙星的降解率最高,Lastre-Acosta等[46]也通过研究发现,在酸性环境下(pH5.5)利用超声法对磺胺嘧啶的降解率较高。
超声法条件温和,对抗生素的降解速度快,无污染,操作方便。但是超声法在抗生素含量较高条件下对抗生素的降解率相对较低[31, 39, 44, 48, 49, 50]。考虑到微生物对抗生素的降解作用,可将超声法与生物法联合应用处理污染废水[53],该联合工艺高效简单清洁,容易操作,应用前景比较好[54]。
1.4 堆肥法由于兽用抗生素的大量使用,使得畜禽粪便里含有大量的抗生素残留,因而未经处理的畜禽粪便直接用于农田,容易造成土壤、作物和地下水的抗生素污染。微生物发酵生产抗生素所产生的药渣因其较高的抗生素残留而被列为工业三废,不仅不能作为农业肥料或工业原料,还会污染环境,影响人体健康,而填埋和焚烧的处理方法费用较高,并且也会造成一定的污染。但是以上所提到的活性污泥法、膜生物反应器法和超声微生物法都是针对含有抗生素的污水处理方法,不适用于含有抗生素的畜禽粪便和药渣等固体废物中抗生素残留的处理,因此堆肥法显示出了自身的优势:既保护了环境,又实现了废弃物的二次利用[55, 56]。堆肥法主要是利用多种微生物的作用,将生物残体、粪便和药渣等进行矿质化、腐殖化和无害化,使各种复杂的有机态养分转化为可溶性养分和腐殖质,同时利用堆积时所产生的高温(60~70 ℃)来杀死原材料中所带的病菌、虫卵和杂草种子等以达到无害化目的。
在堆肥过程中影响抗生素降解率的因素很多,包括堆肥底物、抗生素种类、温度、通气量或通气方式、抗生素浓度和微生物等。不同的底物可能会对抗生素的降解产生不同的影响[57, 58]。Kim等[58]通过实验室堆肥装置试验发现TCs和SAs的降解主要依赖于堆肥底物中添加的木屑;Wu等[59]通过中试规模的猪粪堆肥化使得TCN的降解率为70%;Hu等[60]利用鸡粪、猪粪和水稻秸秆堆肥,使得TCN的降解率达到93%。这可能是不同底物堆肥过程中微生物的生物多样性不同所导致的。为了实现药渣中抗生素的降解,张红娟等[55]设计了林可霉素药渣和牛粪联合堆肥试验,结果显示林可霉素降解率达到99%以上,浸提液种子发芽率从0上升到 70%以上。另外,研究发现药渣堆肥对土壤中微生物增殖的促进作用比一般的牛粪堆肥好,并且药渣堆肥对土壤中微生物的生物多样性没有显著的破坏作用,表明林可霉素菌渣与牛粪的联合堆肥产品已达到无害化和稳定化[56]。
不同的抗生素降解效果在相同的堆肥化条件下会有一定的差异。例如,在同一堆肥条件下,磺胺嘧啶3 d就已全部降解,而TCN 42 d降解率仅为92%;此外,猪粪和木屑按1∶1(V∶V)混合条件下堆肥,磺胺嘧啶3 d完全降解,CTC 21 d完全降解,而环丙沙星56 d仍有17%~31%的残留[61, 62]。
温度会显著影响堆肥对抗生素的降解效果。研究发现,将含有CTC的混合物分别在55 ℃(堆肥温度)和25 ℃温育后堆肥,前者的降解率能达到99%,比后者的降解率高一倍以上。这表明55 ℃比较适合抗生素降解微生物的生存,能够较好地发挥抗生素降解作用[63]。
不同的通气量或通气方式也会影响堆肥法对抗生素的降解效果。Pan等[64]研究了堆肥过程中四种不同的曝气方式(自然通风、翻堆、机械通气和翻堆与机械通气)对抗生素降解的影响。结果表明,翻堆与机械通气并用与其他方式相比能够提高堆肥温度(63 ℃)和延长最高温度的持续时间(4 d,60 ℃)。这可能是由于抗生素的降解主要发生在升温阶段和高温持续阶段的缘故。
在堆肥过程中引入外来有益菌种可以加速抗生素降解。Zhang 等[65]发现在堆肥过程中加入BM菌有利于TCN、CTC和OTC的降解;秦莉等[66]通过在堆肥过程中加入具有降解纤维素和CTC双重功能的复合菌系研究其对CTC的降解作用,结果表明该复合菌系能够在50 ℃快速繁殖,适用于高温好氧堆肥环境,使得CTC的降解率达到82%,与不接复合菌系的处理相比提高60%。
此外,不同的抗生素浓度也会影响堆肥化效果,一般高浓度的抗生素会推迟腐熟时间,因为抗生素浓度越高,对初始的微生物菌群影响越大[60]。
从以下几方面改进堆肥条件可以提高堆肥法对抗生素的降解效果:(1)优化堆肥底物成分配比;(2)针对不同的抗生素设定不同长度的堆肥时间;(3)将堆肥底物先经过高温温育,再进行堆肥;(4)优化通气条件;(5)筛选能够降解抗生素的菌株,尤其是耐高温的菌株,以适应高温堆肥条件。
2 抗生素的微生物降解 2.1 降解条件和效果抗生素特异性降解菌的筛选是利用微生物法降解抗生素最重要的部分。研究发现,真菌和细菌均有可能参与抗生素的降解,目前对抗生素降解菌的筛选鉴定以及降解条件优化情况如表 1所示。
 |
微生物作用下抗生素的降解比较复杂,是微生物在特定环境下通过新陈代谢产生酶等物质,直接或者间接修饰改变抗生素的结构从而使其失活的过程。微生物降解抗生素机制的研究主要包括两个方面:一方面是测定降解过程中微生物的代谢产物,通过对微生物代谢组学、基因组学和蛋白质组学的研究来确定微生物对抗生素的降解机理;另一方面是通过对抗生素降解过程中相关降解产物的连续测定,从而推断抗生素结构的连续性变化规律,即降解途径的研究。
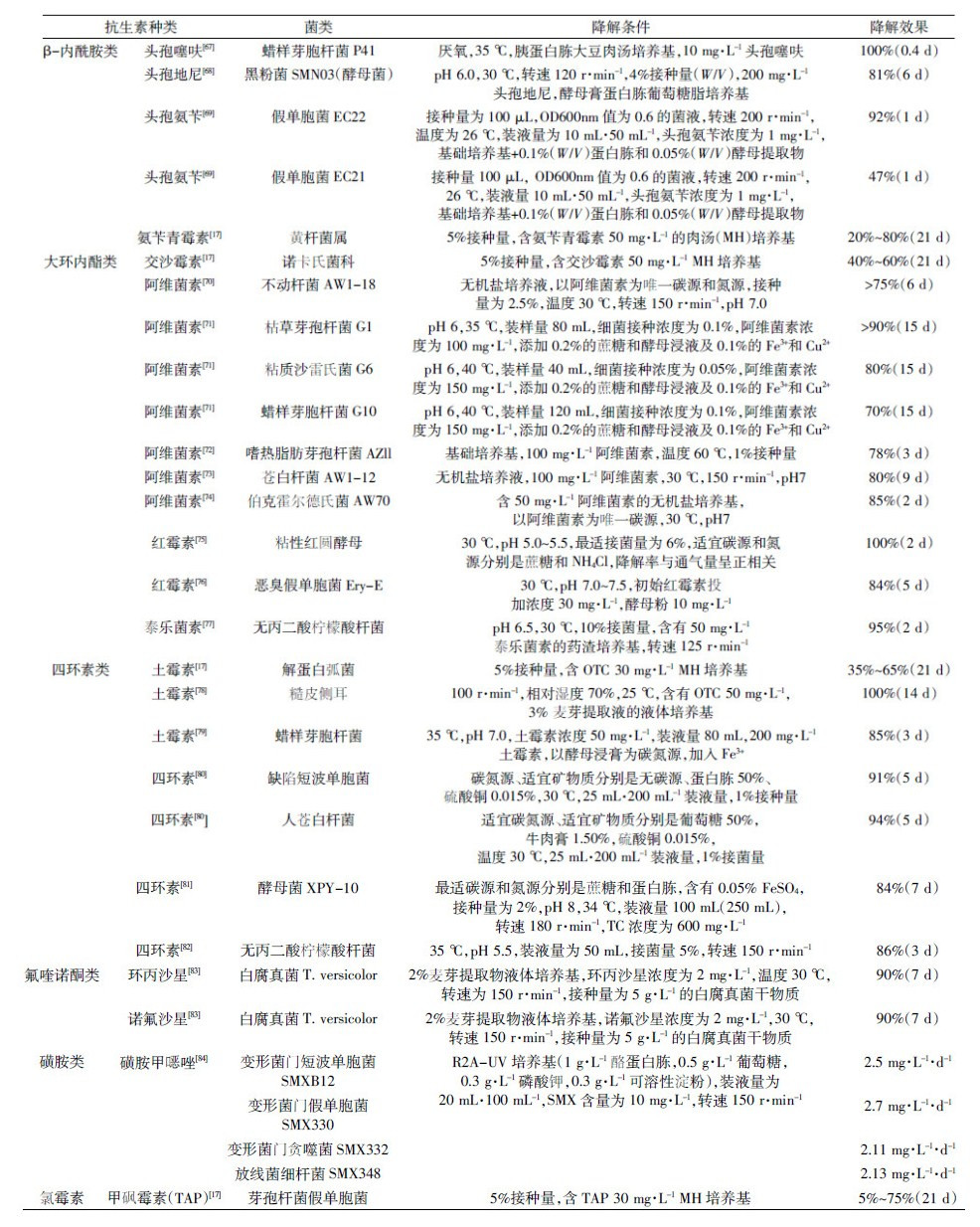
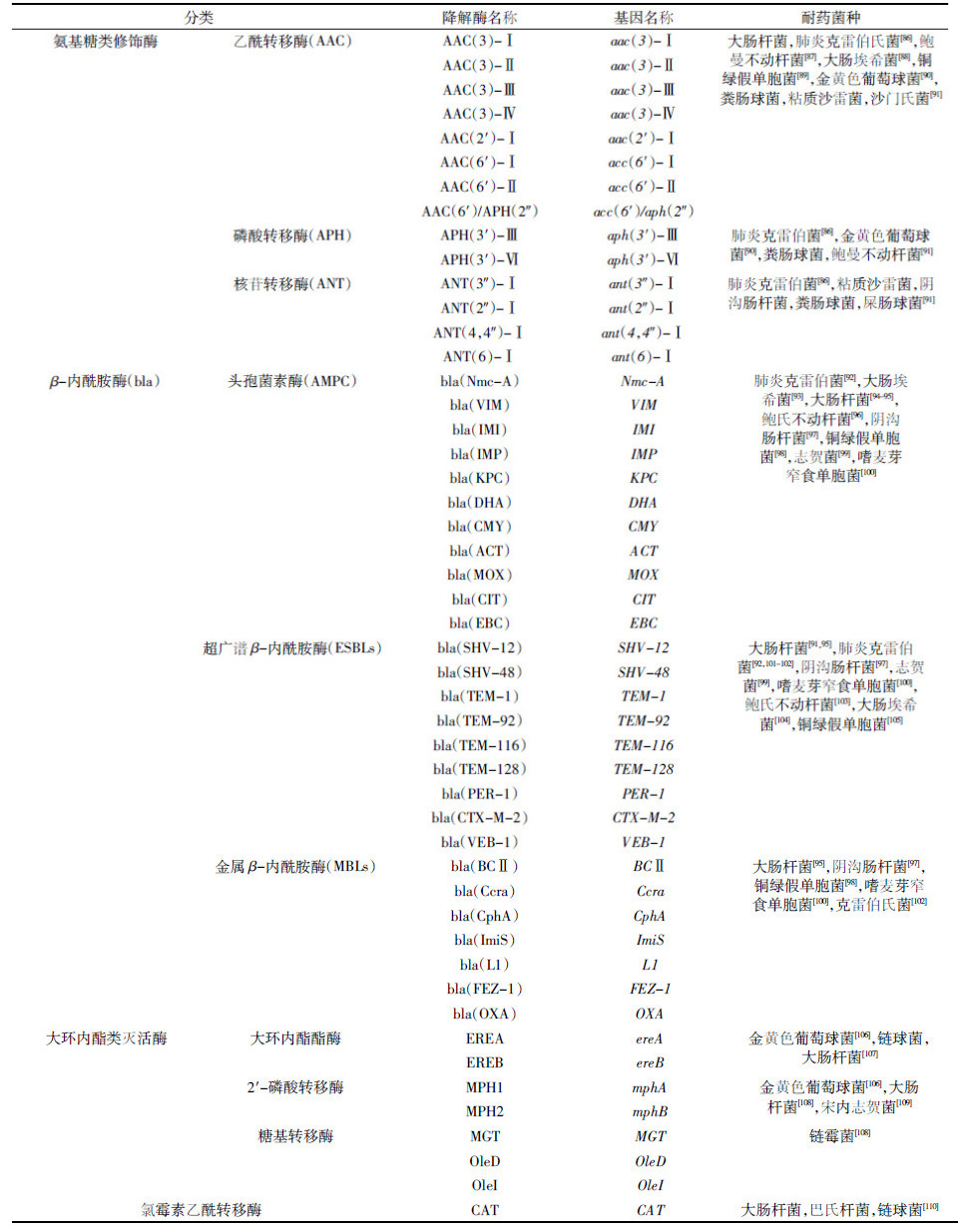
2.2.1 降解酶对于抗生素的微生物降解,其中具有降解功能的主要是抗生素的耐药菌,究其原因是因为这些耐药菌能够产生相应的降解酶,这些酶类进一步通过修饰或水解作用破坏抗生素的分子结构而导致抗生素降解[85]。研究发现抗生素降解酶主要包括以下四大类:β-内酰胺酶、氨基糖苷类修饰酶、大环内酯类钝化酶和氯霉素灭活酶(表 2)。但是以上主要是针对细菌抗生素耐药性的研究,并没有对这些降解酶的抗生素降解条件及其降解效果进行进一步试验。相关报道虽然也有以降解为目的而筛选了一些具有降解抗生素能力的细菌,但是并没有对其降解酶的降解条件进行下一步研究[70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82]。相对于细菌而言,近年来对于具有抗生素降解能力的真菌,包括真菌菌种筛选及其相应降解酶的降解特性和条件等均有一定的研究报道(表 3)。
 |
 |
降解途径作为降解机制研究的重要组成部分,对降解产物的无害化处理起着非常重要的作用。
氨基糖苷类修饰酶主要通过修饰氨基糖苷类抗生素的氨基和羟基等官能团来使抗生素失活。目前发现的氨基糖苷类修饰酶比较多[91],对酶的作用点了解的比较透彻,但是对具体的降解产物了解较少。图 1所示为氨基糖苷类修饰酶对庆大霉素和卡那霉素主要的作用位点[91]。
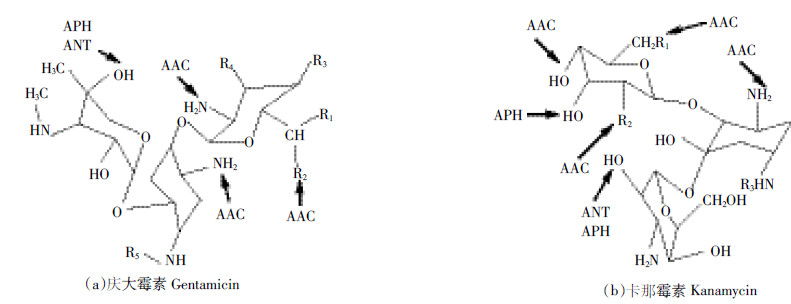
Prieto等[83]在研究影响白腐真菌降解环丙沙星(CIP)和诺氟沙星(NOR)的酶类以及这两种氟喹诺酮类抗生素的降解途径的过程中发现,氟喹诺酮类抗生素在微生物降解酶作用下主要存在三种降解途径:(Ⅰ)哌嗪取代基的氧化;(Ⅱ)单羟基化;(Ⅲ)形成二聚体。如图 2a所示,CIP哌嗪取代基上去掉了C2H2而形成了Cip-1;Cip-1哌嗪取代基中的C2H4N被CH4N取代,形成Cip-2;Cip-3在接种白腐真菌3 d后出现,并且很快被代谢掉,这可能是发生了开哌嗪环而形成Cip-4;第3 d还检测出了Cip-5和Cip-6,这两种产物都是CIP通过C-C共价作用形成,之后又会发生哌嗪基团的断裂、环丙基的去除和羟基化等代谢作用。在最终的培养基中只检测到了Cip-2、Cip-4和Cip-5,所以白腐真菌对CIP矿化可能还存在其他途径。如图 2b所示,接种白腐真菌1 d后NOR开哌嗪环,在氨基部位添加了羧酸而形成Nor-3,经2~3 d Nor-3哌嗪取代基上去掉了C2H2而转化成Nor-1,之后Nor-1哌嗪取代基中的C2H4N被NH2取代形成Nor-2。
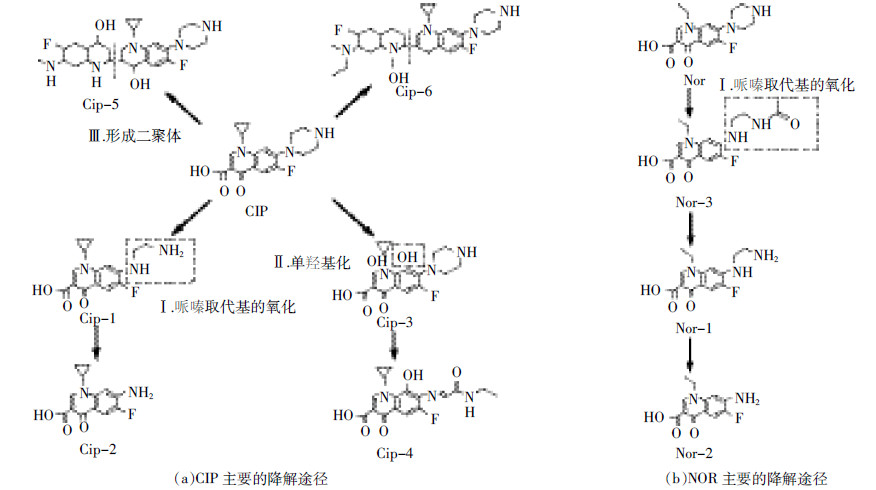
对于头孢类抗生素的微生物降解机理研究表明,在头孢类的β-内酰胺类抗生素的微生物降解中糠酸基团侧链的断裂,即杂环硫醇侧链C3位置的消除是其降解开始时的一个主要步骤,β-内酰胺环的开环是其再降解的一个主要步骤(图 3)[67, 68]。例如,在分析蜡样芽胞杆菌P41对头孢噻呋、头孢曲松钠和头孢泊肟降解途径过程中,发现这三种抗生素最主要的代谢产物都是硫代糠酸基团,该基团是β-内酰胺酶水解后从C3位置被消除所得[63]。
Migliore等[78]利用糙皮侧耳菌在实验室条件下实现了四环素类抗生素OTC的降解,并通过质谱分析发现该菌通过菌丝吸收OTC后再进行降解,推测OTC中的酰胺基转化为乙酰基而成为2-乙酰基-2-去酰胺土霉素(ADOTC),该种产物比OTC的抗菌性低,具有较高的亲油性,毒性相对较低(图 4)。
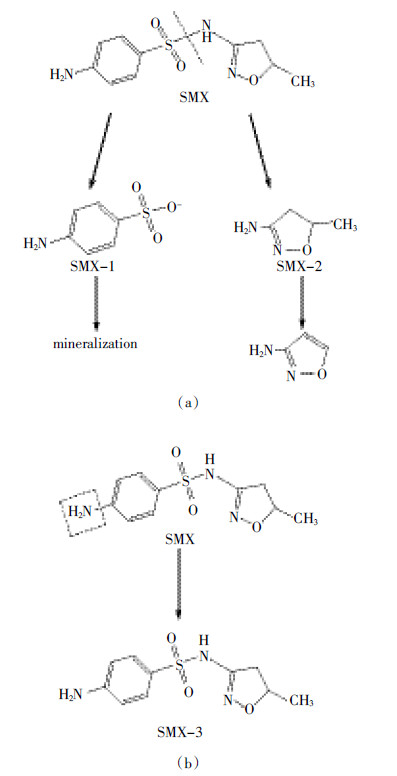
磺胺类抗生素SMX在常温好氧避光条件下可以作为唯一碳源和氮源或者共代谢基质而被活性污泥中两种微生物群落降解[27]。当SMX作为共代谢基质而被异养微生物降解时,其主要产物是3-氨基-5-甲基-异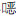 唑(SMX-1)和磺化4-苯胺(SMX-2),其中前者比较稳定,而后者会继续矿化(图 5a)。当SMX作为唯一的碳源和氮源时,除了以上两种产物外还因氨基被羟基取代而生成羟基-N-(5-甲基-1,2-
唑(SMX-1)和磺化4-苯胺(SMX-2),其中前者比较稳定,而后者会继续矿化(图 5a)。当SMX作为唯一的碳源和氮源时,除了以上两种产物外还因氨基被羟基取代而生成羟基-N-(5-甲基-1,2-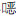 唑-2-yl)苯-1-磺胺(SMX-3)(图b)。
唑-2-yl)苯-1-磺胺(SMX-3)(图b)。
在研究大环内酯类抗生素泰乐菌素的微生物降解机制过程中,发现泰乐菌素降解酶的作用位点不是泰乐菌素的糖苷键和共轭体系,且起降解作用的酶是胞内酶[113]。
总之,微生物对抗生素的降解比较复杂,尤其是不同种类抗生素由于结构不同,微生物降解途径会差异很大,概括起来微生物对抗生素的降解途径主要包括羟基化/去羟基化作用、取代基的氧化作用、裂合作用、取代作用、水解作用和基团转移作用等。
3 展望(1)优化微生物降解抗生素组合工艺,有效利用污泥中的微生物种群资源,提高堆肥效率。
(2)研究开发新型高效的污水和废渣处理设备。
(3)针对不同的抗生素筛选降解能力强的特异性菌株,或者通过诱导驯化,再筛选出能够降解多种抗生素的菌株。
(4)对微生物降解抗生素的机理进行深入研究,从而促进降解菌的无害化。
(5)由于降解菌的筛选可能在一定程度上导致耐药基因的扩散,降解菌的试验要尽量在实验室条件下进行,并且加速对降解酶制剂的研制和生产应用。
(6)在抗生素降解的基础上,进一步加强对降解产物毒性和再降解的后续研究,最终达到抗生素及其降解产物整体的无害化处理。
| [1] | Cheng D M, Liu X H, Wang L, et al. Seasonal variation and sediment-water exchange of antibiotics in a shallower large lake in North China[J]. Science of the Total Environment, 2014, 476:266-275. |
| [2] | Zhou L J, Ying G G, Liu S, et al. Occurrence and fate of eleven classes of antibiotics in two typical wastewater treatment plants in South China[J]. Science of the Total Environment, 2013, 452:365-376. |
| [3] | Kumar K, Gupta S C, Chander Y, et al. Antibiotic use in agriculture and its impact on the terrestrial environment[J]. Advances in Agronomy, 2005, 87:1-54. |
| [4] | Zhao H, Zhou J L, Zhang J. Tidal impact on the dynamic behavior of dissolved pharmaceuticals in the Yangtze Estuary, China[J]. Science of the Total Environment, 2015, 536:946-954. |
| [5] | Ahmed M, Rajapaksha A, Lim J, et al. Distribution and accumulative pattern of tetracyclines and sulfonamides in edible vegetables of cucumber, tomato, and lettuce[J]. Journal of Agricultural and Food Chemistry, 2015, 63(2):398-405. |
| [6] | Matongo S, Birungi G, Moodley B, et al. Pharmaceutical residues in water and sediment of Msunduzi River, KwaZulu-Natal, South Africa[J]. Chemosphere, 2015, 134:133-140. |
| [7] | Pennacchio A, Varriale A, Esposito G, et al. A rapid and sensitive assay for the detection of benzylpenicillin(PenG) in milk[J]. Plos One, 2015, 10(7). doi:10.1371/journal.pone.0132396. |
| [8] | Sun L R, Yu W T, Ma Q, et al. Quantitative analysis of amoxicillin, its major metabolites and ampicillin in eggs by liquid chromatography combined with electrospray ionization tandem mass spectrometry[J]. Food Chemistry, 2016, 192:313-318. |
| [9] | Crane M, Watts C, Boudard T. Chronic aquatic environmental risks from exposure to human pharmaceuticals[J]. Science of the Total Environment, 2006, 367(1):23-41. |
| [10] | 张浩, 罗义, 周启星. 四环素类抗生素生态毒性研究进展[J]. 农业环境科学学报, 2008, 27(2):407-413. ZHANG Hao, LUO Yi, ZHOU Qi-xing. Research advancement of eco-toxicity of tetracycline antibiotics[J]. Journal of Agro-Environment Science, 2008, 27(2):407-413. |
| [11] | Xu Y G, Yu W T, Ma Q, et al. Occurrence of(fluoro)quinolones and(fluoro)quinolone resistance in soil receiving swine manure for 11 years[J]. Science of the Total Environment, 2015, 530:191-197. |
| [12] | Fraqueza, Maria J. Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages[J]. International Journal of Food Microbiology, 2015, 212:76-88. |
| [13] | 世界卫生组织. 2014年全球抗生素耐药性评估报告[R]. 世界卫生组织. 2015:103-108. World Health Organization. Antimicrobial resistance:global report on surveillance 2014[R]. World Health Organism, 2015:103-108. |
| [14] | 范卫红. 纳滤处理水中抗生素的研究[D]. 北京:北京化工大学, 2012. FAN Wei-hong. The research of antibiotics disposal in water by nanofiltration process[D]. Beijing:Beijing University of Chemical technology, 2012. |
| [15] | 陈建发, 刘福权. 微电解-生物滤池等耦合处理抗生素类混合工业废水[J]. 现代化工, 2014, 34(2):120-125. CHEN Jian-fa, LIU Fu-quan. Treatment of antibiotics-based mixed industrial wastewater by microelectrolysis coupled biofilter[J]. Modern Chemical Industry, 2014, 34(2):120-125. |
| [16] | 罗玉, 黄斌, 金玉, 等. 污水中抗生素的处理方法研究进展[J]. 化工进展, 2014, 33(9):2471-2477. LUO Yu, HUANG Bin, JIN Yu, et al. Research progress in the degradation of antibiotics wastewater treatment[J]. Chemical Industry and Engineering Progress, 2014, 33(9):2471-2477. |
| [17] | Teruya M, Hiroshi H, Hiroyuki K, et al. Bacterial degradation of antibiotic residues in marine fish farm sediments of Uranouchi Bay and phylogenetic analysis of antibiotic-degrading bacteria using 16S rDNA sequences[J]. Fisheries Science, 2006, 72(4):811-820. |
| [18] | Prado N, Ochoa J, Amrane A, et al. Biodegradation and biosorption of tetracycline and tylosin antibiotics in activated sludge system[J]. Process Biochemistry, 2009, 44(11):1302-1306. |
| [19] | Kim S, Eichhorn P, Jensen J N, et al. Removal of antibiotics in wastewater:Effect of hydraulic and solid retention times on the fate of tetracycline in the activated sludge process[J]. Environmental Science & Technology, 2005, 39(15):5816-5823. |
| [20] | 李慧. 四环素类抗生素(TCs)在活性污泥系统中的去除特性研究[D]. 泰安:山东农业大学, 2013. LI Hui. The study of removal of tetracycline antibiotics(TCs) on activated sludge[D]. Taian:Shandong Agricultural University, 2013. |
| [21] | Li B, Zhang T. Biodegradation and adsorption of antibiotics in the activated sludge process[J]. Environmental Science & Technology, 2010, 44(9):3468-3473. |
| [22] | Anke G, Christa S M, Adriano J, et al. Fate of sulfonamides, macrolides, and trimethoprim in different wastewater treatment technologies[J]. Science of the Total Environment, 2007, 372(2-3):361-371. |
| [23] | Huang M H, Tian S X, Chen D H, et al. Removal of sulfamethazine antibiotics by aerobic sludge and an isolated Achromobacter sp. S-3[J]. Journal of Environmental Sciences, 2012, 24(9):1594-1599. |
| [24] | Ingerslev F, Sorensen B H. Biodegradability properties of sulfonamides in activated sludge[J]. Environmental Toxicology and Chemistry, 2000, 19(10):2467-2473. |
| [25] | Yang S F, Lin C F, Lin Y C, et al. Sorption and biodegradation of sulfonamide antibiotics by activated sludge:Experimental assessment using batch data obtained under aerobic conditions[J]. Water Research, 2011, 45(11):3389-3397. |
| [26] | Yang S F, Lin C F, Lin Y C, et al. Fate of sulfonamide antibiotics in contact with activated sludge:Sorption and biodegradation[J]. Water Research, 2012, 46(4):1301-1308. |
| [27] | Müller E, Schussler W, Horn H, et al. Aerobic biodegradation of the sulfonamide antibiotic sulfamethoxazole by activated sludge applied as co-substrate and sole carbon and nitrogen source[J]. Chemosphere, 2013, 92(8):969-978. |
| [28] | Xu J, Sheng G P, Ma Y, et al. Roles of extracellular polymeric substances(EPS) in the migration and removal of sulfamethazine in activated sludge system[J]. Water Research, 2013, 47(14):5298-5306. |
| [29] | García N D, Gómez A Z, Navalón A, et al. Removal and degradation characteristics of quinolone antibiotics inlaboratory-scale activated sludge reactors under aerobic, nitrifying and anoxic conditions[J]. Journal of Environmental Management, 2013, 120:75-83. |
| [30] | Guo R X, Xie X D, Chen J Q. The degradation of antibiotic amoxicillin in the Fenton-activated sludge combined system[J]. Environmental Technology, 2015, 36(7):844-851. |
| [31] | 杜兆林, 郑彤, 刘丽艳, 等. 污水处理厂中抗生素去除情况研究[C]//持久性有机污染物论坛:暨第六届持久性有机污染物全国学术研讨会论文集. 2011:244-246. DU Zhao-lin, ZHENG Tong, LIU Li-yan, et al. Analysis of antibiotics removal in sewage treatment plants[C]. 2011:244-246. |
| [32] | 周静博. 膜生物反应器(MBR)对青霉素废水处理的实验研究[D]. 天津:天津工业大学, 2008. ZHOU Jing-bo. Experiment research about penicillin waste water disposal by MBR[D]. Tianjin:Tianjin Polytechnic University, 2008. |
| [33] | Qiu G L, Song Y H, Zeng P, et al. Characterization of bacterial communities in hybrid upflow anaerobic sludge blanket(UASB)-membrane bioreactor(MBR) process for berberine antibiotic wastewater treatment[J]. Bioresource Technology, 2013, 142:52-62. |
| [34] | Sahar E, Messalem R, Cikurel H, et al. Fate of antibiotics in activated sludge followed by ultrafiltration(CAS-UF) and in a membrane bioreactor(MBR)[J]. Water Research, 2011, 45(16):4827-4836. |
| [35] | Shen L, Yuan X, Shen W H, et al. Positive impact of biofilm on reducing the permeation of ampicillin through membrane for membrane bioreactor[J]. Chemosphere, 2014, 97:34-39. |
| [36] | 蒋岚岚, 张万里, 杨薇兰, 等. MBR工艺处理污水效果及影响因素分析[J]. 中国给排水, 2012, 28(24):49-52. JIANG Lan-lan, ZHANG Wan-li, YANG Wei-lan, et al. Analysis on treeatment effect and influence factors of MBR process[J]. China Water and Wastewater, 2012, 28(24):49-52. |
| [37] | Garcia Galan M J, Diaz-Cruz M S, Barcelo D. Removal of sulfonamide antibiotics upon conventional activated sludge and advanced membrane bioreactor treatment[J]. Analytical and Bioanalytical Chemistry, 2012, 404(5):1505-1515. |
| [38] | Garcia N D, Gomez A Z, Navalon A, et al. Removal of quinolone antibiotics from wastewaters by sorption and biological degradation in laboratory-scale membrane bioreactors[J]. Science of the Total Environment, 2013, 442:317-328. |
| [39] | 靖丹枫, 贾仁勇, 白新征, 等. 固定化膜生物反应器处理含抗生素污水[J]. 环境工程学报, 2012, 6(5):1495-1499. JING Dan-feng, JIA Ren-yong, BAI Xin-zheng, et al. Treatment of sewage containing antibiotics by immobilized membrane bioreactor[J]. Techniques and Equipment for Environmental Pollution Control, 2012, 6(5):1495-1499. |
| [40] | Xia S Q, Ren Y, Feng F, et al. Effect of solids retention time on antibiotics removal performance and microbia communities in an A/O-MBR process[J]. Bioresource Techology, 2012, 106:36-43. |
| [41] | 扬程, 郭劲松, Belinda S M, 等. 污泥龄对活性污泥处理微量磺胺类药物的影响[J]. 重庆大学学报, 2012, 35(6):63-71. YANG Cheng, GUO Jin-song, Belinda S M, et al. Impact of sludge residence time on the relative biodegradation and biosorption of sulfonamide antibiotics in activated sludge[J]. Journal of Chongqing University, 2012, 35(6):63-71. |
| [42] | Wang J X, Li Kun, Wei Y S, et al. Performance and fate of organics in a pilot MBR-NF for treating antibiotic production wastewater with recycling NF concentrate[J]. Chemosphere, 2015, 121:92-100. |
| [43] | Dolar D, Gros M, Rodriguez M S, et al. Removal of emerging contaminants from municipal wastewater with an integrated membrane system MBR-RO[J]. Journal of Hazardous Materials, 2012, 239:64-69. |
| [44] | 郭照冰, 周飞, 张超智, 等. 超声降解水中的磺胺嘧啶[J]. 环境工程学报, 2012, 6(9):2143-2147. GUO Zhao-bing, ZHOU Fei, ZHANG Chao-zhi, et al. Degradation of sulfadiazine by ultrasonic[J]. Techniques and Equipment for Environmental Pollution Control, 2012, 6(9):2143-2147. |
| [45] | 李再兴, 剧盼盼, 左剑恶, 等. 微波强化Fenton氧化法深度处理抗生素废水研究[J]. 工业水处理, 2012, 32(6):52-56. LI Zai-xing, JU Pan-pan, ZUO Jian-e, et al. Study on the advanced treatment of antibiotic wastewater by microwave-assisted Fenton oxidation[J]. Industrial Water Treatment, 2012, 32(6):52-56. |
| [46] | Lastre-Acosta A M, Cruz-Gonzalez G, Nuevas-Paz L, et al. Ultrasonic degradation of sulfadiazine in aqueous solutions[J]. Environment Science Pollution Resource, 2015, 22(2):918-925. |
| [47] | Wei H, Li J, Li K B, et al. Degradation of levofloxacin by sonolysis-assisted H2O2 in aqueous solution[J]. China Environmental Science, 2013, 33(2):257-262. |
| [48] | Hoseinia M, Safaria G H, Kamani H, et al. Sonocatalytic degradation of tetracycline antibiotic in aqueous solution by sonocatalysis[J]. Toxicological and Environmental Chemistry, 2013, 95(10):1680-1689. |
| [49] | Wang X K, Wang Y N, Li D L, et al. Degradation of tetracycline in water by ultrasonic irradiation[J]. Water Science and Techology, 2013, 67(4):715-721. |
| [50] | Wei H, Li J, Gao Y, et al. Enhancement of chloromethane on ultrasonic degradation of levofloxacin[J]. Journal of Northwest A & F University-Natural Science, 2013, 41(3):147-152. |
| [51] | Wang Y, Zhang H, Chen L. Ultrasound enhanced catalytic ozonation of tetracycline in a rectangular air-lift reactor[J]. Catalysis Today, 2011, 175(1):283-292. |
| [52] | 郭喜丰, 肖广全, 马丽莉, 等. 超声波降解四环素类抗生素废水[J]. 环境工程学报, 2014, 8(4):1503-1509. GUO Xi-feng, XIAO Guang-quan, MA Li-li, et al. Ultrasonic degradation of tetracyclines in aqueous solution[J]. Chinese Journal of Environmental Engineering, 2014, 8(4):1503-1509. |
| [53] | 马丽莉. 超声波-SBR处理含抗生素、激素猪场废水研究[D]. 重庆:西南大学, 2012. MA Li-li. Study on piggery wastewater contained antibiotics and hormone treated by Ultrasonication-SBR[D]. Chongqing:Southwest University, 2012. |
| [54] | 林海龙, 宋鸽, 司亮, 等. 抗生素废水生物处理法的研究进展[J]. 中国农学通报, 2012, 28(11):258-261. LIN Hai-long, SONG Ge, SI Liang, et al. Advances in study on the biological treatment of antibiotic wastewater[J]. Chinese Agricultural Science Bulletin, 2012, 28(11):258-261. |
| [55] | 张红娟, 郭夏丽, 王岩. 林可霉素菌渣与牛粪联合堆肥实验研究[J]. 环境工程学报, 2011, 5(1):231-235. ZHANG Hong-juan, GUO Xia-li, WANG Yan. Study on co-composting of lincomycin fermentation dregs and cattle manure[J]. Chinese Journal of Environmental Engineering, 2011, 5(1):231-235. |
| [56] | 张红娟. 抗生素菌渣堆肥化处理研究[D]. 郑州:郑州大学, 2010. ZHANG Hong-juan. Research of the composting treatment of antibiotic mushroom dregs[D]. Zhengzhou:Zhengzhou University, 2010. |
| [57] | Mitchell S M, Ullman J L, Bary A, et al. Antibiotic degradation during thermophilic composting[J]. Water Air Soil Pollution, 2015, 226(2):13. |
| [58] | Kim K R, Owens G, Ok Y S, et al. Decline in extractable antibiotics in manure-based composts during composting[J]. Waste Management, 2012, 32(1):110-116. |
| [59] | Wu X F, Wei Y S, Zheng J X, et al. The behavior of tetracyclines and their degradation products during swine manure composting[J]. Bioresource Technology, 2011, 102(10):5924-5931. |
| [60] | Hu Z H, Liu Y L, Chen G W, et al. Characterization of organic matter degradation during composting of manure-straw mixtures spiked with tetracyclines[J]. Bioresource Technology, 2011, 102(15):7329-7334. |
| [61] | Selvam A, Zhao Z Y, Li Y C, et al. Degradation of tetracycline and sulfadiazine during continuous thermophilic composting of pig manure and sawdust[J]. Environmental Technology, 2013, 34(16):2433-2441. |
| [62] | Selvam A, Zhao Z Y, Wong J W C. Composting of swine manure spiked with sulfadiazine, chlortetracycline and ciprofloxacin[J]. Bioresource Technology, 2012, 126:412-417. |
| [63] | Arikan O A, Mulbry W, Rice C. Management of antibiotic residues from agricultural sources:Use of composting to reduce chlortetracycline residues in beef manure from treated animals[J]. Journal of Hazardous Materials, 2009, 164(2-3):483-489. |
| [64] | Pan X, Qiang Z M, Ben W W. Effects of high-temperature composting on degradation of antibiotics in swine manure[J]. Journal of Ecology and Rural Environment, 2013, 29(1):64-69. |
| [65] | Zhang S Q, Zhang F D, Liu X M, et al. Degradation of antibiotics and passivation of heavy metals during thermophilic composting process[J]. Scientia Agricultura Sinica, 2006, 39(2):337-343. |
| [66] | 秦莉, 高茹英, 李国学, 等. 外源复合菌系对堆肥纤维素和金霉素降解效果的研究[J]. 农业环境科学学报, 2009, 28(4):820-823. QIN Li, GAO Ru-ying, LI Guo-xue, et al. Decomposition effect of additive of composite microbial system on cellulose and chlortetracycline in composting[J]. Journal of Agro-Environment Science, 2009, 28(4); 820-823. |
| [67] | Erickson B D, Elkins C A, Mulli L B, et al. A metallo-beta-lactamase is responsible for the degradation of ceftiofur by the bovine intestinal bacterium Bacillus cereus P41[J]. Veterinary Microbiology, 2014, 172(3/4):499-504. |
| [68] | Selvi A, Salam J A, Das N. Biodegradation of cefdinir by a novel yeast strain, Ustilago sp. SMN03 isolated from pharmaceutical wastewater[J]. World J Microbiol Biotechnol, 2014, 30(11):2839-2850. |
| [69] | Lin B K, Liu J L, Liu X J, et al. Characterization of cefalexin degradation capabilities of two Pseudomonas strains isolated from activated sludge[J]. Journal of Hazardous Materials, 2015, 282:158-164. |
| [70] | 胡秀虹, 李景壮, 叶胜蓝, 等. 一株阿维菌素降解菌AW1-18的筛选与分类鉴定[J]. 生物技术通报, 2012, 10:223-228. HU Xiu-hong, LI Jing-zhuang, YE Sheng-lan, et al. Screening and identification of avermectin-degrading bacterium strain AW1-18[J]. Biotechnology Bulletin, 2012, 10:223-228. |
| [71] | 闫彩虹. 三株阿维菌素高效降解菌的筛选、鉴定及降解特性研究[D]. 扬州:扬州大学, 2011. YAN Cai-hong. Screening, Identification and degradation characteristic research of three avermectin-degrading bacterium strains[D]. Yangzhou:Yangzhou University, 2011. |
| [72] | 魏艳丽, 李纪顺, 扈进冬, 等. 降解阿维菌素耐高温菌株AZ11的分离及降解特性[J]. 山东科学, 2013, 26(4):16-19. WEI Yan-li, LI Ji-shun, HU Jin-dong, et al. Solation and degradation characteristics of an avermectin-degrading and thermophilic bacterial strain AZ11[J]. Shandong Science, 2013, 26(4):16-19. |
| [73] | 胡秀虹, 黄剑. 阿维菌素降解菌AW1-12的筛选与分类鉴定[J]. 西南农业学报, 2013, 26(2):583-586. HU Xiu-hong, HUANG Jian. Screening and identification of avermectin-degrading bacterium strain AW1-12[J]. Southwest China Journal of A gricultural Sciences, 2013, 26(2):583-586. |
| [74] | 李荣, 管晓进, 陈荣宗, 等. 阿维菌素降解菌株AW70的分离鉴定及降解特性研究[J]. 土壤, 2009, 41(4):607-611. LI Rong, GUAN Xiao-jin, CHEN Rong-zong, et al. Isolation and identification of an avermectins-degrading strain AW70 and its degrading characteristics[J]. Soils, 2009, 41(4):607-611. |
| [75] | 许晓玲. 红霉素、四环素降解菌的筛选、鉴定及其降解性能研究[D]. 杭州:浙江大学, 2008. XU Xiao-ling. Screening and identification of erythromycin-degrading strains and tetracycline-degrading strains and their degradation characteristics[D]. Hangzhou:Zhejiang University, 2008. |
| [76] | 毛菲菲, 刘畅, 何梦琦, 等. 红霉素降解菌的筛分及其降解特性的研究[J]. 环境科学与技术, 2013, 36(7):9-12. MAO Fei-fei, LIU Chang, HE Meng-qi, et al. Isolation and identification of an erythromycin degradation bacterium and study on its biodegradation characteristics[J]. Environmental Science & Technology, 2013, 36(7):9-12. |
| [77] | 刘力嘉, 谢丽, 张作义, 等. 泰乐菌素高效降解菌的筛选及降解特性研究[J]. 农业环境科学学报, 2011, 30(5):1027-1030. LIU Li-jia, XIE Li, ZHANG Zuo-yi, et al. Isolation and degradation characteristics of a tylosin-degrading strain[J]. Journal of Agro-Environment Science, 2011, 30(5):1027-1030. |
| [78] | Migliorea L, Fiori M, Spadonia A, et al. Biodegradation of oxytetracycline by Pleurotus ostreatus mycelium:A mycoremediation technique[J]. Journal of Hazardous Materials, 2012, 215-216:227-232. |
| [79] | 王志强, 张长青, 王维新. 土霉素降解菌的筛选及其降解特性研究[J]. 中国兽医科学, 2011, 41(5):536-540. WANG Zhi-qiang, ZHANG Chang-qing, WANG Wei-xin. Screening and characterization of an oxytetracycline degrading bacterium[J]. Chinese Veterinary Science, 2011, 41(5):536-540. |
| [80] | 许晓玲, 李卫芬, 雷剑, 等. 四环素降解菌的选育、鉴定及其降解特性[J]. 农业生物技术学报, 2011, 19(3):549-556. XU Xiao-ling, LI Wei-fen, LEI Jian, et al. Screening, identification and degradation characteristics of tetracycline-degrading strains[J]. Journal of Agricultural Biotechnology, 2011, 19(3):549-556. |
| [81] | 冯福鑫, 许旭萍, 程群星, 等. 四环素高效降解酵母菌Trichosporon mycotoxinivorans XPY-10降解特性[J]. 环境工程学报, 2013, 7(12):4779-4785. FENG Fu-xin, XU Xu-ping, CHENG Qun-xing, et al. Degradation characteristics of tetracycline hydrochloride by Trichosporon mycotoxinivorans XPY-10[J]. Chinese Journal of Environmental Engineering, 2013, 7(12):4779-4785. |
| [82] | 马志强, 马玉龙, 谢丽, 等. 微生物法降解药渣中残留四环素的试验研究[J]. 环境科学与技术, 2012, 35(1):46-49. MA Zhi-qiang, MA Yu-long, XIE Li, et al. Experimental study on microbial degradation of tetracycline residues in antibiotic waste[J]. Environmental Science & Technology, 2012, 35(1):46-49. |
| [83] | Prieto A, Moder M, Rodil R, et al. Degradation of the antibiotics norfloxacin and ciprofloxacin by a white-rot fungus and identification of degradation products[J]. Bioresource Technology, 2011, 102(23):10987-10995. |
| [84] | Herzog B, Lemmer H, Horn H, et al. Characterization of pure cultures isolated from sulfamethoxazole-acclimated activated sludge with respect to taxonomic identification and sulfamethoxazole biodegradation potential[J]. BMC Microbiology, 2013,12. doi:10.1186/1471-2180-13-276. |
| [85] | 陈如登, 吴波浪. 抗生素耐药性耐药机制的探讨[J]. 福建畜牧兽医, 2006, 28(1):59-60. CHEN Ru-deng, WU Bo-lang. Discussion about antibiotic resistance mechanisms[J]. Fujian Journal of Animal Husbandry and Veterinary, 2006, 28(1):59-60. |
| [86] | 糜祖煌, 黄支密, 秦玲. 鲍氏不动杆菌耐药性和氨基糖苷类修饰酶、β-内酰胺酶基因研究[J]. 中华医院感染学杂志, 2004, 14(9):968-971. MI Zu-huang, HUANG Zhi-mi, QIN Ling. Resistance and genotyping of aminoglycosides modifying enzymes and β-Lactamases in acinetobacter baumannii[J]. Chinese Journal of Nosocomiology, 2004, 14(9):968-971. |
| [87] | Liu Z R, Ling B D, Zhou Li M. Prevalence of 16S rRNA methylase, modifying enzyme, and extended-spectrum beta-lactamase genes among Acinetobacter baumannii isolates[J]. Journal of Chemotherapy, 2015, 27(4):207-212. |
| [88] | Dias G V, Bohrer L F, Oliveira F B, et al. Detection and characterization of multidrug-resistant enterobacteria bearing aminoglycoside-modifying gene in a university hospital at Rio de Janeiro, Brazil, along three decades[J]. Biomedica, 2015, 35(1):117-124. |
| [89] | Michalska A D, Anna D, Sacha P T, et al. Prevalence of resistance to aminoglycosides and fluoroquinolones among Pseudomonas aeruginosa strains in a University Hospital in Northeastern Poland[J]. Brazilian Journal of Microbiology, 2014, 45(4):1455-1458. |
| [90] | Mohammadi S, Sekawi Z, Monjezi A, et al. Emergence of SCCmec type Ⅲ with variable antimicrobial resistance profiles and spa types among methicillin-resistant Staphylococcus aureus isolated from healthcare- and community-acquired infections in the west of Iran[J]. International Journal of Infectious Diseases, 2014, 25:152-158. |
| [91] | Van de Klundert J A M, Vliegenthart J S. PCR detection of genes coding for aminoglycoside-modifying enzymes[M]//Persing D H, Smith T F, Tenover F C, et al. Diagnostic Molecular Microbiology, American Society for Microbiology. Washington DC, 1993:547-552. |
| [92] | YoonY K, Cheong H W, Pai H, et al. Molecular analysis of a prolonged spread of Klebsiella pneumoniae co-producing DHA-1 and SHV-12β-lactamases[J]. Journal of Microbiology, 2011, 49(3):363-368. |
| [93] | 董方, 徐樨巍, 宋文琪. 临床分离大肠埃希菌和克雷伯菌质粒介导AmpC β-内酰胺酶的研究[J]. 中华医学杂志, 2010, 90(38):2723-2725. DONG Fang, XU Xi-wei, SONG Wen-qi. Prevalence of plasmid-mediated AmpC beta-lactamases in Escherichia coli and Klebsieila spp. isolated in children[J]. National Medical Journal of China, 2010, 90(38):2723-2725. |
| [94] | Yoo J S, Byeon J, Yang J, et al. High prevalence of extended-spectrum-beta-lactamases and plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae isolated from longterm care facilities in Korea[J]. Diagnostic Microbiology and Infectious Disease, 2010, 67(3):261-265. |
| [95] | Rezaei F, Kalantar D, Delfani S, et al. Characterization co-existence of AmpC, MBLs, TEM and SHV type of beta-lactamases in clinical strains of Escherichia coli and Klebsiella pneumoniae isolated from hospitals of Khorramabad, Iran[J]. Tropical Medicine and International Health, 2015, 20(Suppl1):295-295. |
| [96] | Liu Y R, Liu X Q. Detection of AmpC beta-lactamases in Acinetobacter baumannii in the Xuzhou region and analysis of drug resistance[J]. Experimental and Therapeutic Medicine, 2015, 10(3):933-936. |
| [97] | 黄支密, 诸葛青云, 糜祖煌. 阴沟肠杆菌I类整合酶基因及质粒AmpC酶基因检测[J]. 江西医学检验, 2005, 23(1):21-24. HUANG Zhi-mi, ZHUGE Qing-yun, MI Zu-huang. Detection of the class 1 integrase gene and plasmid-AmpC enzyme genes in Enterobacter cloacae[J]. Jiangxi Journal of Medical Laboratory Sciences, 2005, 23(1):21-24. |
| [98] | 黄义山, 龙彦, 方莉, 等. 多药耐药铜绿假单胞菌金属β-内酰胺酶及相关基因检测[J]. 中国临床医师杂质, 2010, 4(9):1623-1627. HUANG Yi-shan, LONG Yan, FANG Li, et al. Study on the resistant genes of metal β-lactamase(MBLs) of the multidrug resistant mechanism of Pseudomonas aeruginosa[J]. Chinese Journal of Clinicians, 2010, 4(9):1623-1627. |
| [99] | 张传领, 葛玉梅, 沈丽芳, 等. 同时产ESBLS和AMPC酶志贺菌临床菌株的分离及鉴定[J]. 中华微生物学和免疫学杂质, 2014, 34(4):251-255. ZHANG Chuan-ling, GE Yu-mei, SHEN Li-fang, et al. Characteristics of clinical Shigella isolates producing the extended-spectrum and AmpC β-lactamases[J]. Chinese Journal of Microbiology Immunology, 2014, 34(4):251-255. |
| [100] | 罗卉丽, 陈姗姗, 杨宏伟, 等. 35株多重耐药嗜麦芽窄食单胞菌的β-内酰胺酶耐药基因分布研究[J]. 检验医学与临床, 2015, 10(12):2839-2844. LUO Hui-li, CHEN Shan-shan, YANG Hong-wei, et al. Study of distribution of β-lactamase drug resistance gene in 35 strains of multi-drug resistant Stenotrophomonas malto-philia[J]. Clinical Chemistry and Laboratory Medicine, 2015, 10(12):2839-2844. |
| [101] | Bora A, Hazarika N K, Shukla Sanket K, et al. Prevalence of bla(TEM), bla(SHV) and bla(CTX-M) genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India[J]. Indian Journal of Pathology and Microbiology, 2014, 57(2):249-254. |
| [102] | Lob S H, Biedenbach D J, Badal R E, et al. Antimicrobial resistance and resistance mechanisms of Enterobacteriaceae in ICU and non-ICU wards in Europe and North America:SMART 2011-2013[J]. Journal of Global Antimicrobial Resistance, 2015, 3(3):190-197. |
| [103] | Alyamani E J, Khiyami M A, Booq R Y, et al. Molecular characterization of extended-spectrum beta-lactamases(ESBLs) produced by clinical isolates of Acinetobacter baumannii in Saudi Arabia[J]. Annals of Clinical Microbiology and Antimicrobials, 2015, 14:38. |
| [104] | Tseng S P, Wang S F, Cheng Y, et al. Characterization of fosfomycin resistant extended-spectrum beta-lactamase-producing Escherichia coli isolates from human and pig in Taiwan[J]. Plos One, 2015, 10(8). doi:10.1371/journal.pone.0135864 |
| [105] | Oliver A, Mulet X, Lopez C C, et al. The increasing threat of Pseudo-monas aeruginosa high-risk clones[J]. Drug Resistance Updates, 2015, 21-22:41-59. |
| [106] | 黄烈, 林广城, 张银辉, 等. 葡萄球菌对大环内酯类抗生素耐药及诱导耐药表型与基因型的相关性研究[J]. 国际检验医学杂志, 2009, 30(6):551-554. HUANG Lie, LIN Guang-cheng, ZHANG Yin-hui, et al. The correlation between phenotype and genotype of macrolides resistance in staphylococci[J]. International of Journal Laboratory Medicine, 2009, 30(6):551-554. |
| [107] | 潘丽萍. 大环内酯类抗生素耐药的流行状况和机制[J]. 现代医学, 2008, 36(5):370-372. PAN Li-ping. The popularity of macrolide antibiotics resistant condition and mechanism[J]. Modern Medical Journal, 2008, 36(5):370-372. |
| [108] | 曾焱华, 吴移谋. 细菌对大环内酯类抗生素耐药的机制及控制策略[J]. 国外医学, 2003, 26(4):18-22. ZENG Yan-hua, WU Yi-mou. Macrolide antibiotics resistant mechanism and the control strategy[J]. Foreign Medical Sciences, 2003, 26(4):18-22. |
| [109] | Howie R L, Folster J P, Bowen A, et al. Reduced azithromycin susceptibility in shigella sonnei, United States[J]. Microbial Drug Resistance, 2010, 16(4):245-248. |
| [110] | 杜向党, 阎若潜, 沈建忠. 氯霉素类药物耐药机制的研究进展[J]. 动物医学进展, 2004, 25(2):29-31. DU Xiang-dang, YAN Ruo-qian, SHEN Jian-zhong. The research progress on mechanism of resistance of phenicolss[J]. Progress in Veterinary Medicine, 2004, 25(2):29-31. |
| [111] | Wen X H, Jia Y N, Li J X, et al. Degradation of tetracycline and oxytetracycline by crude lignin peroxidase prepared from Phanerochaete chrysosporium:A white rot fungus[J]. Chemosphere, 2009, 75(8):1003-1007. |
| [112] | Wen X H, Jia Y N, Li J X, et al. Enzymatic degradation of tetracycline and oxytetracycline by crude manganese peroxidase prepared from Phanerochaete chrysosporium[J]. Journal of Hazardous Materials, 2010, 177(1-3):924-928. |
| [113] | 王艳. 药渣中残留泰乐菌素的微生物降解途径及其降解产物研究[D]. 宁夏:宁夏大学, 2013. WANG Yan. A study on microbial degradation pathway and products of tylosin residues in antibiotic waste[D]. Ningxia:Ningxia University, 2013. |
 2016, Vol. 35
2016, Vol. 35