2. 南开大学环境科学与工程学院, 环境污染过程与基准教育部重点实验室/天津市城市生态环境修复与污染防治重点实验室, 天津 300350
2. Key Laboratory of Pollution Processes and Environmental Criteria(Ministry of Education)/Tianjin Key Laboratory of Environmental Remediation and Pollution Control, College of Environmental Science and Engineering, Nankai University, Tianjin 300350, China
我国是抗生素生产和使用大国,总量一直居于全球首位[1]。一直以来,抗生素被广泛应用于畜禽养殖业中的动物疾病预防和治疗[2-3]。据专家调查和推算,中国每年生产抗生素原料大约21万t,其中9.7万t应用于畜禽养殖业中,占年产抗生素总量的46.1%。进入到动物体内的抗生素只有少部分能被代谢、吸收和降解,大部分残留在动物粪便中[4-7],对肠道微生物造成选择性压力,动物体内存在的抗生素抗性细菌(Antibiotic resistant bacteria,ARBs)和抗生素抗性基因(Antibiotic resistance genes,ARGs),不仅极大阻碍畜禽养殖粪便的有效资源化利用,更可能给周边生态环境带来潜在的健康风险[8-10]。
畜禽粪便的排放造成大量携带ARGs的抗性细菌进入到水和土壤环境,通过水平接合转移将抗性传递给土著微生物,甚至进入食物链,给生态和人类健康带来严重威胁[11-13]。当前针对畜禽养殖场(鸡、猪、牛和羊等)及周边环境ARGs污染情况的调研已有大量报道[14-19]。例如,Tina等[20]研究比利时畜禽粪便中抗生素和ARGs迁移行为,发现猪粪样品中以四环素类(tetB、tetL、tetM、tetO、tetQ和tetW)、磺胺类(sulⅡ)和大环内酯类(ermB和ermF)抗性基因为主;Haley等[21]调查发现美国17个商业奶牛场养殖粪便ARGs以四环素类和氨基糖苷类最为丰富,其在小牛粪便中的平均丰度(1.8×102)要明显高于其在乳牛粪便中的丰度(1.7×10-1);Qian等[22]在鸡、猪和牛的大型养殖场粪便样品中共检测到109种ARGs,且鸡场和猪场ARGs污染水平显著高于牛场;Kong等[23]调查我国吉林省羊场养殖粪便中ARGs污染特征,利用高通量PCR技术检测到多种抗生素抗性,包括氨基糖苷类[aacC(6)I1、aacC2、aacC4、strA和strB)、β-内酰胺类(blaCTX-M、blaSHV和blaTEM)、四环素类(tetB、tetC、tetE、tetO和tetQ)、大环内酯-林酰胺类-链脲类(mphA、mphB、ermA、ermB和ermC)、磺胺类(sulⅠ和sulⅡ)、喹诺酮类(qnrA和qnrS)、多黏菌素(MCR-1)和新德里金属- β-内酰胺抗性(NDM-1)等,相对丰度为8.99×10−9~3.00×10−1;Liu等[24]甚至在5年不使用抗生素的养鸡场粪便也检测到超过80种的ARGs,其中aadA、aadA1、aadA2、strB、tetM、tetK和tetX的基因丰度均超过10-3,cphA基因丰度高达10-1。最近,谷艳茹等[25]调查天津市家庭农场粪污ARGs赋存特征,发现tetO、tetQ、tetW和ermB含量最为丰富,同时检出到blaOXA-1、blaTEM-1和blaampC等β-内酰胺类抗性基因,这些畜禽粪肥的施用可显著增加土壤环境中ARGs的丰度(约8~18倍)。国内外研究重点集中在畜禽养殖场粪便和周边环境的ARGs来源和污染程度解析,但有关不同规模化养殖场的ARGs污染特征差异还未见报道。
环境中ARGs主要通过接合、转导和转化3种方式进行横向迁移[26-31]。研究表明,有机化合物可通过调控可动遗传因子、引起细胞SOS反应、胁迫细胞形成自然感受态、影响生物膜形成、改变细胞膜通透性、与胞外质粒形成加合物等方式,影响ARGs在微生物之间的水平转移效率;还可通过影响微生物群落结构,改变ARGs的表达水平。例如,Kang等[32]发现多环芳烃(菲、萘等)可通过与pUC19质粒形成非共价键复合物,显著抑制氨苄青霉素抗性基因(Ampr)的转化效率;刘璐[33]发现水体中溶解性有机质腐植酸和富里酸都对sul的增殖和传播具有明显的抑制效果,并且随着浓度的升高抑制作用逐渐增强;Zhao等[34]发现腐植酸(25 mg·L-1)可影响水处理系统中的微生物群落结构,并显著抑制四环素抗性基因(tetC、tetG、tetW和tetX)和Ⅰ类整合子基因(intI)的表达水平;Zhou等[35]研究我国华东地区高污染城市河流内ARGs污染特征,发现总ARGs的绝对丰度与总有机碳和总溶解性有机氮含量呈显著正相关(P < 0.01)。上述研究均表明有机质的含量和成分可一定程度上影响环境介质中ARGs的污染水平。然而,相比于其他影响因子(如pH、温度和抗生素)的大量报道[36-38],畜禽养殖粪便中残留有机质含量与ARGs污染的关系还不清楚。
华北地区(京津冀)作为我国重要的工业基地,其土壤微生物环境和食品安全一直都是被关注的重点。本实验选取天津和河北两地区规模化的养鸡场和养猪场为研究对象,探讨不同规模(大型、中型和小型)的养殖场畜禽粪便中ARGs(四环素类tet、磺胺类sul、大环内酯类erm和喹诺酮类qns)的污染特征;测定粪便中残留有机碳(Organic carbon,OC)和有机氮(Organic carbon,ON)的含量,分析畜禽粪便中残留有机质含量与ARGs污染水平的相关性,以期为控制我国规模化畜禽养殖场中抗生素和ARGs污染与保障土壤环境健康提供科学依据,为环境生态风险评价提供参考。
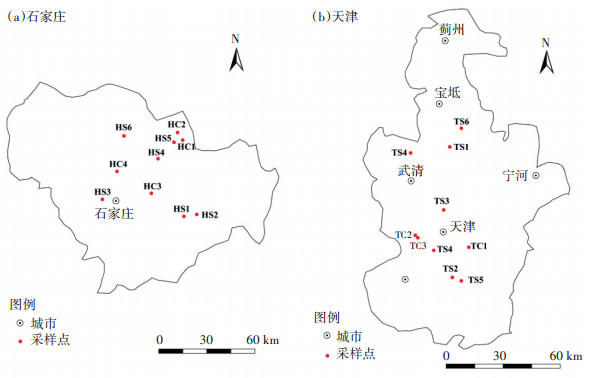
1 材料与方法 1.1 样品的采集、运输与保存选取具有一定规模的养猪场和养鸡场作为研究对象,采样点主要分布在天津市西青区和河北省石家庄市及周边地区,具体采样点分布见图 1。以动物的年出栏量将养殖场规模划分为大型、中型和小型。养鸡场年出栏5万羽以上为中型养鸡场(MC,HC1~HC4、TC1~TC2),年出栏1万到5万羽为小型养鸡场(SC,TC3~TC4);养猪场年出栏10 000头以上为大型养猪场(LS,HS1~HS2、TS1~TS2),5 000~10 000头为中型养猪场(MS,HS3~HS4、TS3~TS4),5 000头以下为小型养猪场(SS,HS5~HS6、TS5~TS6)。采样前对选取的养殖场养殖年限和抗生素使用情况进行调研。中型和大型养殖场养殖年限均在8年以上,小型养殖场养殖年限在3年左右。养鸡场常用药物包括磺胺类(磺胺嘧啶和磺胺甲恶唑)、四环素类(四环素、金霉素和强力霉素)、大环内酯类(吉他霉素)、氟喹诺酮类(环丙沙星和诺氟沙星)和氨基糖苷类(庆大霉素和卡那霉素);养猪场常用药物包括磺胺类(磺胺甲嘧啶和磺胺甲恶唑)、四环素类(四环素、金霉素和土霉素)、大环内酯类(泰乐菌素)和氟喹诺酮类(环丙沙星和恩诺沙星)。采样时利用铲子收集猪舍和鸡舍新鲜出栏未经任何处理的粪便,每个养殖场随机选取3个不同的养殖栏收集一定量粪便,然后迅速搅拌混合均匀作为均一样品。粪便收集完毕后立即低温快递运回实验室,提取DNA之前-20 ℃保存。
|
图 1 养殖场采样点分布 Figure 1 The locations of targeted livestock farms |
粪便基因组DNA提取试剂盒FastDNA® SPIN Kit for Feces购买自美国MP Biomedicals生物医药公司;琼脂糖凝胶DNA回收试剂盒(DP208/209)购买自北京天根生化科技有限公司;异丙醇、乙醇、EDTA、Na2EDTA、琼脂粉、Loading Buffer、溴化乙锭(EB)、NaCl、Tris、IPTG、X-gal、氨苄青霉素、琼脂糖、牛肉膏、蛋白胨、酵母浸粉、E.coli感受态细胞、pMD18-T Simple Vector、2×Taq PCR Green Mix等试剂均购自北京鼎国昌盛生物技术有限公司;实时荧光定量PCR试剂盒则由TAKARA提供。实验用水均为双蒸水。
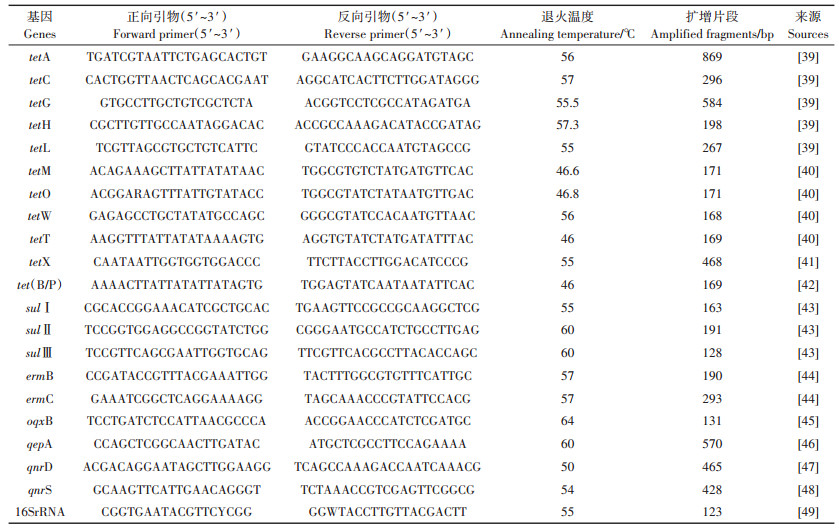
1.3 基因组DNA的提取与PCR反应粪便细菌基因组DNA的提取依照FastDNA® SPIN Kit for Feces试剂盒指示操作进行。DNA提取后,使用微量核酸蛋白测定仪(Nanodrop2000)鉴定核酸浓度及纯度,保证OD260/OD280比值大于1.8,以满足后续PCR实验要求。然后以基因组DNA为模板,经稀释后混合为25 μL PCR反应体系,PCR反应体系为:12.5 μL的2×Taq PCR Green Mix(包含0.1 U·μL-1 Taq DNA Polymerase、2×PCR buffer、3 mmol·L-1 MgCl2和0.4 mmol·L-1 dNTPs)、前端引物0.5 μL(10 mmol· L-1)、后端引物0.5 μL(10 mmol·L-1)、DNA模板1 μL和10.5 μL的双蒸水,空白对照组以双蒸水为模板。检测目标基因包含tet类[tetA、tetC、tetG、tetH、tetL、tetM、tetO、tetW、tetT、tetX和tet(B/P)]、sul类(sulⅠ、sul Ⅱ和sulⅢ)、erm类(ermB和ermC)和qns类(oqxB、qepA、qnrD和qnrS)抗性基因,基因引物序列详情如表 1所示。定性PCR反应程序为:94 ℃预变性3~5 min,94 ℃变性30 s,退火30 s后,72 ℃延伸30 s,共40个循环,最后72 ℃延伸7 min。反应结束后,利用DNA凝胶电泳检测所得条带,统计目标基因的检出率。
|
|
表 1 PCR引物序列 Table 1 The sequence of PCR primers |
选择粪便样品中检出率为100%的目标基因,鉴定ARGs的相对丰度。本实验中ARGs的定量采用绝对定量法,选取16S rRNA作为内参基因。首先绘制标准曲线:目的基因普通PCR后,条带切胶回收,目的基因片段与pMD19-T载体连接,连接产物与50 μL解冻后的E.coli感受态细胞均匀混合,用未加入氨苄青霉素的LB液体培养基适当培养。然后将菌液均匀地涂在含有氨苄青霉素、X-gal和IPTG的LB固体抗性平板上培养,蓝白斑筛选挑选出白色重组菌斑,挑选出的白色菌斑在LB液体培养基上扩大培养后取出抽提质粒,利用Nanodrop2000鉴定所提质粒DNA的浓度及纯度,换算出质粒作为标准品的基因拷贝数,然后将标准质粒梯度稀释,使其浓度在10-7~10-1,经RT-qPCR扩增后作出Ct值与DNA起始浓度有关的线性曲线。通过标准曲线的绘制得到目标基因的扩增曲线公式,目标基因的扩增效率E在84.8%~ 133.9%,相关系数R2基本都保持在0.99以上,线性关系良好。
RT-qPCR反应体系为:12.5 μL TransStart® Top Green qPCR SuperMix、正向和反向引物(10 mmol·L-1)各0.5 μL、DNA模板1 μL和双蒸水10.5 μL。具体反应程序为:94 ℃下预变性5 s,95 ℃变形30 s,退火30 s,72 ℃延伸30 s,总共40个循环,溶解曲线程序在55~95 ℃,每隔0.5 s读数,每个温度停留30 s。程序结束后记录其循环数(Ct值),根据标准曲线计算出起始样品中目标基因的拷贝数,以16S rRNA基因校准,计算得出目标基因在畜禽粪便样品中的相对丰度。
1.5 有机碳和有机氮测定实验采用重铬酸钾氧化-分光光度法(HJ 615— 2011)测定粪便样品中的有机碳(OC)含量,以葡萄糖为标准使用液绘制标准曲线。准确称取0.5~1 g干燥后的粪便样品加入到消解玻璃管中,每个样品做3个平行,然后加入0.5 g硫酸汞和10 mL重铬酸钾溶液,摇匀后缓慢加入7.5 mL浓硫酸,恒定温度135 ℃加热30 min。反应结束后加入约50 mL水冷却至室温,倒入比色管中加水至100 mL,摇匀,静置2 h后取30 mL溶液,2 000 r·min-1离心10 min,静置澄清,收集上清液,在585 nm处以水为参比测定溶液吸光度。干燥粪便样品中的OC含量(质量分数,%)按照如下公式进行计算:
Woc = (A - A0 - a) / (b × m1 × 1000) × 100
式中:Woc为样品中OC的含量(质量分数),mg C·g-1;m1为风干样品质量,g;A为消解液的吸光度;A0为空白组的吸光度;a为标准曲线截距;b为标准曲线斜率。
有机氮(ON)含量由凯氏氮和铵态氮含量差减法计算得出。凯氏氮含量采用全自动凯氏定氮仪(KJELTECTM 8400,FOSS,丹麦)测定,铵态氮含量则采用纳氏试剂比色法(HJ 535—2009)测定。称取干燥粪便样品5 g置于锥形瓶中,加入0.05 mol·L-1的盐酸30 mL,振荡混匀30 min后取出倒入离心管中3 000 r·min-1离心10 min,静置澄清后取上清液5 mL置于容量瓶中定容至100 mL,而后取稀释液2 mL置于50 mL的比色管中,加水至50 mL,然后分别加入1.5 mL的纳氏试剂和1 mL的酒石酸钾钠溶液混匀,在波长420 nm处以水为参比测定吸光度。
1.6 数据分析文中所列数据为3个平行组的平均值,误差棒表示标准偏差。数据的统计和绘制利用IBM SPSS Stastistic 20.0和Origin 9.0软件,显著性使用独立样本t检验进行分析,P < 0.05表示具有显著性差异。利用SIMCA-P 11.5软件进行相关性分析,使用正交偏最小二乘判别(OPLS-DA)方法,VIP(Variable important in projection)是OPLS-DA模型变量的变量权重值,表示有机质对不同种类ARGs的影响强度和解释能力,VIP≥1表示具有显著性。利用Canoco 5.0软件进行典型相关分析(CCA)。
2 结果与讨论 2.1 规模化养殖场粪便中ARGs的检出频率养殖场采集畜禽粪便样品中ARGs的检出频率如表 2所示。实验结果显示,编码外排泵蛋白(Efflux pump proteins,EPP)的tetH和tetL、编码核糖体保护蛋白(Ribosomal protection proteins,RPP)的tetM、tetO和tetW、以及编码酶抑制剂(Enzyme-inhibitor complex,EI)的tetX的检出率均达到100%;对sul基因而言,sul Ⅰ和sulⅡ在所有样品中均有检出,sulⅢ的检出频率则相对较低(85%);两种erm基因在所有养殖场中均检测到;所有qns基因的检出率在80%~95%之间,相比于tet、sul和erm基因,检出率明显偏低。
|
|
表 2 养殖场畜禽粪便中ARGs的检出频率(n=20) Table 2 The detection frequencies of the targeted ARGs in livestock manures (n=20) |
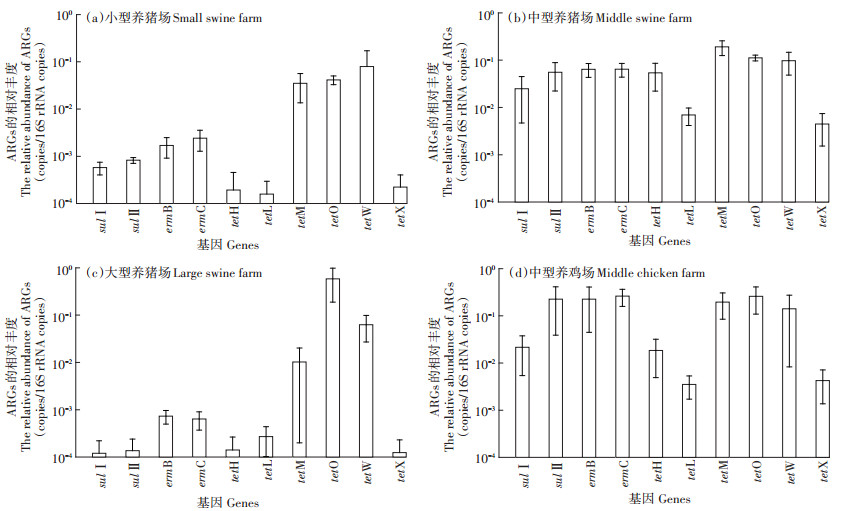
河北和天津地区不同规模的养猪场和养鸡场ARGs相对丰度分别如图 2和图 3所示,结果表明不同种类养殖场粪便中ARGs的污染特征存在显著差异。在猪粪便中,tetM,tetO和tetW总体相对丰度是最高的(4.24×10-3~5.85×10-1),erm基因次之(6.36×10-4~ 6.45×10-2),sul基因的相对丰度最低(1.07×10-4~5.56× 10-2),这种趋势在河北地区尤为明显。而在鸡粪样品中,ARGs整体丰度较高,不同抗生素的ARGs相对丰度相差较小,erm基因(2.51×10-2~2.62×10-1)、sul基因(1.86×10-3~2.26×10-1)与编码RPP的tet基因(1.13× 10-2~2.59×10-1)的相对丰度较为接近,均高于编码EPP(1.31×10-4~1.84×10-2)和EI(8.97×10-4~4.25×10-3)的tet基因相对丰度。
|
图 2 河北地区养殖场畜禽粪便ARGs的相对丰度 Figure 2 The relative abundances of ARGs in manures collected from livestock farms in Hebei Province |
|
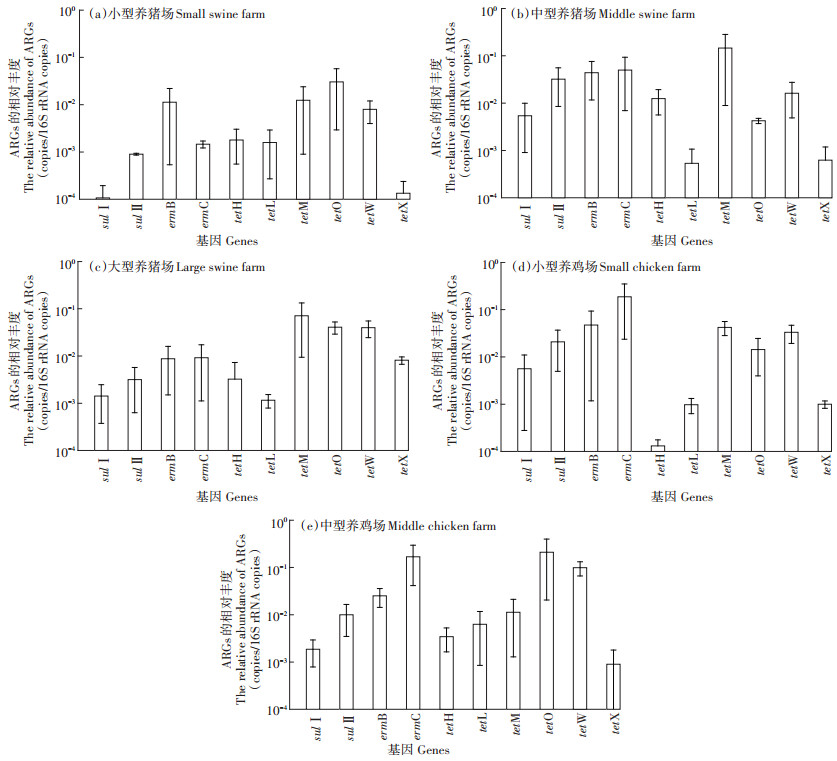
图 3 天津地区养殖场畜禽粪便ARGs的相对丰度 Figure 3 The relative abundances of ARGs in manures collected from livestock farms in Tianjin |
养鸡场粪便中sul和erm基因的相对丰度高于其在养猪场粪便中的相对丰度。例如,河北省鸡粪中sulⅡ基因的相对丰度为2.16×10-2~2.26×10-1,而其在猪粪中的相对丰度则为1.2×10-4~5.56×10-2,研究结果与Cheng等[50]调研我国华东地区畜禽粪便的结论一致。对于畜禽养殖业ARGs污染水平而言,抗生素使用量越高,粪便中残留的抗生素含量越高,ARGs的污染就越严重。Zhao等[51]研究报道指出,我国畜禽养殖业鸡粪中抗生素残留量(1 420.76 mg·g-1)明显高于猪粪。更高含量的抗生素残留量和更严重的选择性压力,导致鸡粪中sul和erm基因的污染水平比猪粪中的更高。意外的是,编码RPP和EPP抗性的tet基因在猪粪和鸡粪中的相对丰度较为接近,这可能与这两类ARGs广泛的宿主菌群和高水平转移扩散能力有关[52]。
ARGs污染水平与其宿主菌的种类、数量以及微生物种属之间的转移能力有关。大多数tet基因坐落于非移动性基因元件上[52],但是编码RPP抗性的tet基因仍然具有较强的转移能力,可通过氧四环素抗性质粒进行水平转移传播[53];而且相对于编码EPP和EI抗性的tet基因,tetM、tetO和tetW具有更广泛的宿主菌(分别为60、24种和12种),导致粪便和环境介质中编码RPP抗性的tet基因相对丰度普遍更高[54]。同样地,实验发现不同抗性机理的tet基因相对丰度趋势为:RPP>EPP>EI,tetM、tetO和tetW的最高相对丰度分别可达1.96×10-1、2.59×10-1和5.85×10-1。此外,大部分erm基因同时编码有大环内酯(Macrolide)、林酰胺类(Lincosamide)和链脲类(Streptogramin)抗性(MLS),可轻易被移动质粒和转座子获得并随之水平接合转移,在环境介质中污染水平也普遍较高[55-56],实验同样发现其相对丰度显著高于编码EPP和EI的tet基因。对于sul基因,sulⅠ和sulⅡ都编码DHPS抗性,且sulⅠ基因多数位于Ⅰ类整合子移动基因元件上[57],大量研究发现环境介质(土壤、水体和污泥等)中sulⅠ基因的相对丰度普遍高于sulⅡ基因[58],但是sulⅡ基因污染水平更易受到人为活动因素干扰[59],这可能是本研究发现畜禽粪便sulⅡ基因相对丰度高于sulⅠ基因的主要原因。重要的是,sulⅡ基因的检测丰度最高可达2.26 × 10-1,明显高于Munir等[60]和Ji等[61]研究动物粪便中检测到的sulⅡ基因丰度,说明近些年随着抗生素的进一步使用,畜禽养殖业中ARGs的污染水平变得更加严重和不可忽视。
2.2.2 不同地区畜禽养殖场粪便样品中ARGs的相对丰度四环素类和磺胺类是畜禽养殖业使用最为频繁的抗病药物,tet和sul两类抗性基因的检出也极为频繁。Heuer等[62]2008年调查发现德国养猪场粪便中sul基因相对丰度的数量级保持在10-5~10-2;2009年检测美国24个畜禽养殖场粪便中ARGs污染水平,结果显示tetW和sulⅠ基因的相对丰度分别在10-3~10-2和10-6~10-5[63];2014年Garder等[64]研究表明erm基因在畜禽粪便样品中难以检测到,但是在排放粪便的周边土壤和水体环境中则检出频率较高。上述研究表明,不同地区畜禽养殖业ARGs的污染水平存在较大差异。在本项研究中,河北地区养猪场和养鸡场ARGs的平均相对丰度分别为4.99×10-2和1.36×10-1,均高于天津市养猪场和养鸡场ARGs的平均相对丰度(分别为1.89×10-2和4.45×10-2),尤其是中型养猪场(P=0.023),表明河北地区畜禽养殖场粪便ARGs污染水平要比天津市更为严重,这可能与当地不同规模的畜禽养殖场抗生素管控政策和措施有关。然而,受限于天津市未采集到中型养鸡场粪便样品和抗生素使用量的不确定,不同地区养殖企业ARGs污染水平差异的具体原因仍需进一步核查。
2.2.3 不同规模的畜禽养殖场粪便中ARGs的相对丰度图 4和表 3结果显示,河北地区小型、中型和大型养猪场粪便ARGs的平均相对丰度为1.62×10-2、6.76× 10-2和6.60×10-2,中型养猪场平均ARGs相对丰度最高,显著高于小型养猪场(P=0.018),而中型与大型养猪场、大型与小型养猪场之间均无显著性差异;天津地区小型、中型和大型养猪场粪便ARGs的平均相对丰度为6.8×10-3、3.12×10-2和1.87×10-2,与河北地区类似,天津中型规模养猪场粪便ARGs丰度显著高于小型养猪场(P=0.039),其他不同规模养猪场之间ARGs污染水平无显著性差异。不同规模养猪场粪便中ARGs的平均相对丰度趋势均为:中型>大型>小型,说明中等规模的养猪场对抗生素的使用更为泛滥,小型养猪场抗生素使用则相对较少。
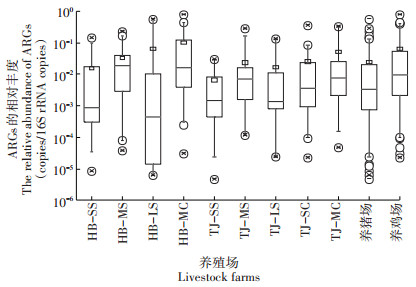
|
图 4 不同规模畜禽养殖场粪便中ARGs的相对丰度 Figure 4 The relative abundance of ARGs in animal manures collected from different scaled livestock farms |
|
|
表 3 河北和天津地区不同规模养殖场粪便中ARGs丰度差异性显著水平(P) Table 3 The statistical significance(P value)of ARGs abundances in different scaled livestock farms in Hebei Province and Tianjin |
河北地区中型养鸡场粪便ARGs的平均相对丰度为1.36×10-1,天津市中型和小型养鸡场粪便ARGs的平均相对丰度分别为5.37×10-2和3.53×10-2,相对于相同地区同等规模的养猪场而言,养鸡场粪便中ARGs相对丰度明显更高;此外,统计学分析结果显示天津市小型和中型养鸡场之间ARGs污染水平无显著性差异(P=0.551)。上述研究结果表明,同一地区鸡类养殖企业对抗生素的使用相比于养猪场可能更为严重,养鸡场无论规模大小,粪便中ARGs的污染水平普遍较高。
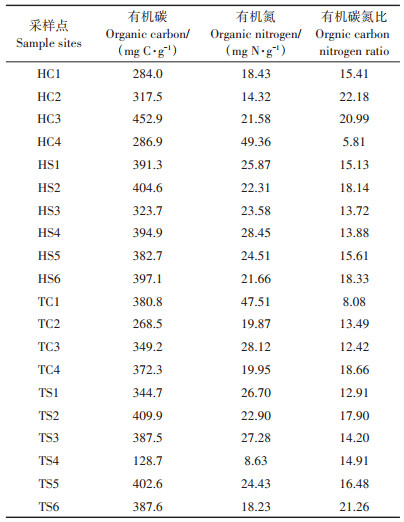
2.3 畜禽粪便中有机质与ARGs污染水平的相关性分析养殖场畜禽粪便中OC和ON的检测含量如表 4所示,除HC4外所有粪便样品中OC/ON比值均超过8。OPLS-DA相关性分析(图 5)结果表明,猪粪中ARGs相对丰度与OC含量的相关性比鸡粪要强,显著相关的基因种类更多(sulⅠ、ermC、tetL、tetM、tetW和tetX),而ARGs丰度与ON和OC/ON的相关性在猪粪和鸡粪中则比较接近。此外,不同种类的ARGs与有机质含量的相关性有差异。tet基因与OC和ON的相关性要明显强于sul和erm基因,其中编码RPP的tet基因与有机质的相关性最强,例如猪粪样品中tetM与OC和OC/ON均显著正相关,tetO与ON和OC/ON均显著相关,tetW则与OC和ON均显著正相关;在鸡粪样品中,tetM和tetW基因则均与ON含量和OC/ON显著相关。
|
|
表 4 天津和河北地区养殖场畜禽粪便中有机碳和有机氮含量 Table 4 The contents of organic carbon and organic nitrogen in collected manures in Tianjin and Hebei Province |
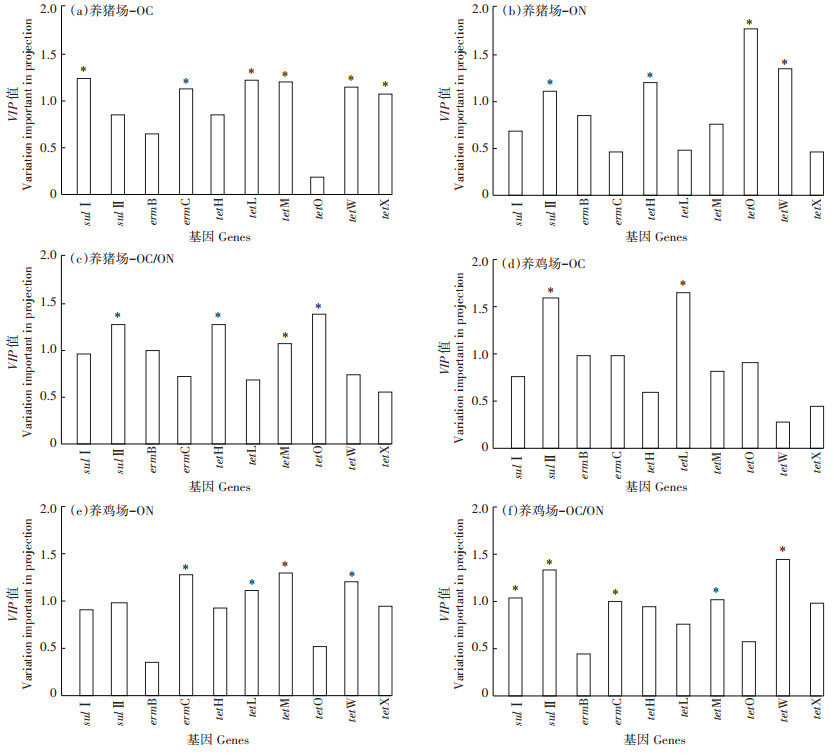
|
*表示具有差异显著性 * represents the statistical significance 图 5 畜禽粪便中ARGs相对丰度与有机质含量OPLS-DA相关分析的变量权重(VIP)值 Figure 5 The statistical VIP value of OPLS-DA analysis between ARGs abundance and organic matter in livestock manures |
已有研究表明,生物可利用碳氮比(C/N)影响着微生物的群落结构及其活性[65],而微生物群落结构与ARGs的丰度和多样性紧密相关[66]。研究发现,鸡粪样品中sulⅠ和sulⅡ基因均与OC/ON呈显著负相关,而在猪粪中则呈正相关;对于tet基因,tetH基因和编码RPP的tet基因相对丰度几乎均与OC/ON呈显著性相关。上述研究结果表明畜禽粪便中OC/ON可能是决定ARGs污染水平的重要因素之一。研究结果还显示ARGs的相对丰度与OC/ON之间的相关性和其与ON之间的相关性几乎完全相反,尤其是显著性相关的基因(sulⅡ、ermC、tetH、tetM、tetO和tetW),表明畜禽养殖场粪便中ON含量与ARGs污染水平有较强的关联性。
畜禽粪便中ARGs污染水平受到多种因子的影响。本研究发现粪便中有机质(尤其是ON)含量与多种ARGs污染水平呈现显著性相关,表明有机质成分和含量可能是调控粪便ARGs赋存特征和水平的重要因素。研究表明,可生物利用的OC和ON等营养物质决定着微生物胞外酶活性和细胞外基质的组分和功能[67],这些胞外酶和胞外基质可通过吸附和水解等作用影响外来分子(包括DNA)转移进入细胞内部的效率和能力。Tets等[68]发现在生物膜中,胞外的蛋白水解酶可显著抑制抗性质粒的接合频率。这些研究结果预示着有机质可能通过影响细菌胞外分泌物的成分和活性,从而影响ARGs在肠道微生物之间的转移和扩散。此外,Hu等[69]发现粪便中OC/ON比值高于8时,反硝化细菌会成为主要的优势菌群,在一定范围内,ON含量越高和有效C/N越大,反硝化效率会增高,反硝化细菌越容易占据优势主导地位;任四伟[70]研究发现养鸡场和养猪场粪便中的单一(62.58%~89.43%和29.62%~49.72%)和多重抗生素类群(95.29%和88.55%)主要是拟杆菌门(Bacteroidetes);蒋志云等[71]采用Miseq高通量测序技术发现,在污水处理反硝化过程中,优势微生物菌群始终以变形菌门(Proteobacteria)和拟杆菌门(Bacteroidetes)为主。综合上述研究结果,有机质(OC和ON)可能通过调控畜禽粪便中的优势抗性菌群及其胞外分泌物的成分和活性,影响畜禽养殖业粪便中ARGs的污染水平。
CCA分析结果(图 6)表明:sul基因与OC/ON显著相关,tetM、tetO和tetW均具有丰富的宿主菌群[28],更容易受到有机质含量的影响,与OC和ON均表现出非常强的相关性,这与OPLS-DA相关性分析的结果是一致的。相比之下,erm基因和tetX基因与有机质含量的相关性很弱。众多研究表明erm基因和编码EI的tet基因在养殖业粪便和周边土壤环境中普遍存在且丰度较高[72-73],表明畜禽养殖业ARGs污染水平的环境影响因子仍需进一步探索和研究。
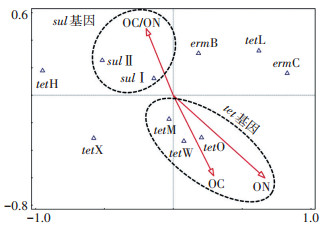
|
图 6 畜禽粪便中ARGs丰度与有机质含量的典型对应分析 Figure 6 Canonical correspondence analysis of ARGs abundance and the contents of organic matter in livestock manures |
(1)华北地区养鸡场粪便中sul和erm基因的相对丰度显著高于其在猪粪中的水平,编码RPP的tet基因(10-2~10-1)在两种畜禽粪便中的相对丰度比较接近,高于sul(10-4~10-1)、erm(10-3~10-1)、编码EPP和编码EI的tet基因(10-5~10-2)的相对丰度。
(2)不同规模养猪场粪便中ARGs污染水平趋势为:中型>大型>小型,不同规模的养鸡场ARGs的平均相对丰度无显著差异。
(3)养殖粪便中ARGs污染水平与有机质含量高度相关。tetM、tetO和tetW基因与有机碳(OC)和有机氮(ON)含量高度相关(VIP>1),sul基因则与OC/ON呈明显的相关性,ON和生物可利用C/N是影响畜禽养殖粪便ARGs污染水平的重要因素。
| [1] |
李红娜, 阿旺次仁, 李斌绪, 等. 兽用抗生素研究的文献计量学分析[J]. 农业环境科学学报, 2017, 36(11): 2297-2306. LI Hong-na, A Wang-ci-ren, LI Bin-xu, et al. A bibliometric assessment of research into antibiotics in poultry and livestock breeding[J]. Journal of Agro-Environment Science, 2017, 36(11): 2297-2306. |
| [2] |
Saemah A K, Meyer M T, Boxall A B, et al. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics(VAs)in the environment[J]. Chemosphere, 2006, 65(5): 725-759. DOI:10.1016/j.chemosphere.2006.03.026 |
| [3] |
Mathew A G, Cissell R, Liamthong S. Antibiotic resistance in bacteria associated with food animals:A united states perspective of livestock production[J]. Foodborne Pathogens and Disease, 2007, 4(2): 115-133. DOI:10.1089/fpd.2006.0066 |
| [4] |
Menz J, Olsson O, Kümmerer K. Antibiotic residues in livestock manure:Does the EU risk assessment sufficiently protect against microbial toxicity and selection of resistant bacteria in the environment?[J]. Journal of Hazardous Materials, 2019, 379(5): 120807. |
| [5] |
李厚禹, 邵振鲁, 李碧菡, 等. 畜禽环境中抗生素的去除及其风险评估[J]. 生态毒理学报, 2020, 15(1): 79-93. LI Hou-yu, SHAO Zhen-lu, LI Bi-han, et al. The removal and risk assessment of antibiotics in livestock environment[J]. Asian Journal of Ecotoxicology, 2020, 15(1): 79-93. |
| [6] |
Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields[J]. Current Opinion in Microbiology, 2011, 14(3): 236-243. DOI:10.1016/j.mib.2011.04.009 |
| [7] |
Hu X G, Zhou Q X, Luo Y. Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, Northern China[J]. Environmental Pollution, 2010, 158(9): 2992-2998. DOI:10.1016/j.envpol.2010.05.023 |
| [8] |
Shea K M. Antibiotic resistance:What is the impact of agricultural uses of antibiotics on children's health?[J]. Pediatrics, 2003, 112: 253-258. |
| [9] |
罗义, 周启星. 抗生素抗性基因(ARGs)——一种新型环境污染物[J]. 环境科学学报, 2008, 28(8): 1499-1505. LUO Yi, ZHOU Qi-xing. Antibiotic resistance genes(ARGs)as emerging pollutants[J]. Acta Scientiae Circumstantiae, 2008, 28(8): 1499-1505. |
| [10] |
邹威, 罗义, 周启星. 畜禽粪便中抗生素抗性基因(ARGs)污染问题及环境调控[J]. 农业环境科学学报, 2014, 33(12): 2281-2287. ZOU Wei, LUO Yi, ZHOU Qi-xing. Pollution and environmental regulation of antibiotic resistance genes(ARGs)in livestock manure[J]. Journal of Agro-Environment Science, 2014, 33(12): 2281-2287. |
| [11] |
Zhou S Y D, Zhu D, Giles M, et al. Does reduced usage of antibiotics in livestock production mitigate the spread of antibiotic resistance in soil, earthworm guts, and the phyllosphere?[J]. Environment International, 2020, 136: 105359. DOI:10.1016/j.envint.2019.105359 |
| [12] |
Mclnnes R S, MaCallum G E, Lamberte L E, et al. Horizontal transfer of antibiotic resistance genes in the human gut microbiome[J]. Current Opinion in Microbiology, 2020, 53: 35-43. DOI:10.1016/j.mib.2020.02.002 |
| [13] |
Cheng J H, Tang X Y, Cui J F. Effect of long-term manure slurry application on the occurrence of antibiotic resistance genes in arable purple soil(entisol)[J]. Science of the Total Environment, 2019, 647(10): 853-861. |
| [14] |
Pu Q, Zhao L, Li Y, et al. Manure fertilization increase antibiotic resistance in soils from typical greenhouse vegetable production bases, China[J]. Journal of Hazardous Materials, 2020, 391(5): 122267. |
| [15] |
Sen L, Qin Y, Liu J, et al. Profiles of antibiotic resistome with animal manure application in black soils of Northeast China[J]. Journal of Hazardous Materials, 2020, 384(15): 121216. |
| [16] |
Wang L, Wang J, Wang J, et al. Distribution characteristics of antibiotic resistant bacteria and genes in fresh and composted manures of livestock farms[J]. Science of the Total Environment, 2019, 695(10): 133781. |
| [17] |
杨亦文, 米见对, 邹永德, 等. 广东省规模化猪场猪粪中典型耐药基因污染分析[J]. 中国兽医学报, 2018, 38(8): 1559-1567. YANG Yi-wen, MI Jian-dui, ZOU Yong-de, et al. Analysis of typical antibiotic resistance genes in the manure of large scale swine farms in Guangdong Province[J]. Chinese Journal of Veterinary Science, 2018, 38(8): 1559-1567. |
| [18] |
何良英.典型畜禽养殖环境中抗生素耐药基因的污染特征与扩散机理研究[D].广州: 中国科学院研究所院(广州地球化学研究所), 2016. HE Liang-ying. Contamination and spread of antibiotic resistance genes in the environment of representative livestock feedlots[D]. Guangzhou: Guangzhou Institute of Geochemistry, Chinese Academy of Sciences, 2016. |
| [19] |
Yuan Q, Zhai Y, Mao B, et al. Antibiotic resistance genes and int I1 prevalence in a swine wastewater treatment plant and correlation with metal resistance, bacterial community and wastewater parameters[J]. Ecotoxicology and Environmental Safety, 2018, 161(15): 251-259. |
| [20] |
Tina V, Geertrui R, Freddy H, et al. Presence and fate of antibiotic residues, antibiotic resistance genes and zoonotic bacteria during biological swine manure treatment[J]. Ecotoxicology and Environmental Safety, 2019, 175(15): 29-38. |
| [21] |
Haley B J, Kim S W, Salaheen S, et al. Differences in the microbial community and resistome structures of feces from preweaned calves and lactating dairy cows in commercial dairy herds[J]. Foodborne Pathogens and Disease, 2020, 17: 7. |
| [22] |
Qian X, Gu J, Sun W, et al. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting[J]. Journal of Hazardous Materials, 2018, 344(15): 716-722. |
| [23] |
Kong L, Wang B, Wang Y, et al. Characterization of bacterial community changes and antibiotic resistance genes in lamb manure of different incidence[J]. Scientific Reports, 2019, 9: 10101. DOI:10.1038/s41598-019-46604-y |
| [24] |
Liu Y, Michael D, Marc M, et al. Antibiotic resistance genes in antibiotic-free chicken farms[J]. Antibiotics, 2020, 9(3): 120. |
| [25] |
谷艳茹, 韩秉君, 黄继元, 等. 天津市家庭农场养殖粪污耐药基因赋存特征及风险评估[J]. 农业环境科学学报, 2020, 39(2): 394-402. GU Yan-ru, HAN Bing-jun, HUANG Ji-yuan, et al. Occurrence characteristics and risk assessment of resistance genes in livestock waste from family farms in Tianjin City, China[J]. Journal of Agro-Environment Science, 2020, 39(2): 394-402. |
| [26] |
D'Costa V M, McGrann K M, Hughes D W, et al. Sampling the antibiotic resistome[J]. Science, 2006, 311(5759): 374-377. DOI:10.1126/science.1120800 |
| [27] |
Wright G D. The antibiotic resistome:The nexus of chemical and genetic diversity[J]. Nature Reviews Microbiology, 2007, 5(3): 175-186. DOI:10.1038/nrmicro1614 |
| [28] |
Webber M A, Piddock L J V. The importance of efflux pumps in bacterial antibiotic resistance[J]. Journal of Antimicrobial Chemotherapy, 2003, 51(1): 9-11. DOI:10.1093/jac/dkg050 |
| [29] |
Salyers A A, Shoemaker N B, Stevens A M, et al. Conjugative transposons:An unusual and diverse set of integrated gene transfer elements[J]. Microbiological Reviews, 1995, 59(4): 579-590. DOI:10.1128/MMBR.59.4.579-590.1995 |
| [30] |
Whittle G, Shoemaker N B, Salyers A A. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT[J]. Journal of Bacteriology, 2002, 184(14): 3839-3847. DOI:10.1128/JB.184.14.3839-3847.2002 |
| [31] |
Ghosh D, Roy K, Williamson K E, et al. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA[J]. Applied and Environmental Microbiology, 2008, 74(2): 495-502. |
| [32] |
Kang F, Hu X, Liu J, et al. Noncovalent binding of polycyclic aromatic hydrocarbons with genetic bases reducing the in vitro lateral transfer of antibiotic resistant genes[J]. Environmental Science & Technology, 2015, 49: 10340-10348. |
| [33] |
刘璐.水环境因子对磺胺类抗生素抗性基因的影响[D].大连: 大连海洋大学, 2018. LIU Lu. The influence of water quality on sulfonamide antibiotic resistance gene[D]. Dalian: Dalian Ocean University, 2018. |
| [34] |
Zhao J, Li Y, Li Y, et al. Application of humic acid changes the microbial communities and inhibits the expression of tetracycline resistance genes in 4-chlorophenol wastewater treatment[J]. Journal of Environmental Management, 2019, 250(15): 109463. |
| [35] |
Zhou Z, Zheng J, Wei Y, et al. Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters[J]. Environmental Science and Pollution Research, 2017, 24(30): 23753-23762. DOI:10.1007/s11356-017-0032-0 |
| [36] |
隋倩雯, 张俊亚, 魏源送, 等. 畜禽养殖过程抗生素使用与耐药病原菌及其抗性基因赋存的研究进展[J]. 生态毒理学报, 2015, 10(5): 20-34. SUI Qian-wen, ZHANG Jun-ya, WEI Yuan-song, et al. Veterinary antibiotics use, occurrence of antibiotic resistance pathogen and its antibiotic resistance genes in animal production:An overview[J]. Asian Journal of Ecotoxicology, 2015, 10(5): 20-34. |
| [37] |
Li M, Ray P, Knowlton K F, et al. Fate of pirlimycin and antibiotic resistance genes in dairy manure slurries in response to temperature and pH adjustment[J]. Science of the Total Environment, 2020, 710(25): 136310. |
| [38] |
Zhou B, Wang C, Zhao Q, et al. Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms[J]. Journal of Hazardous Materials, 2016, 320(15): 10-17. |
| [39] |
Aminov R, Chee-Sanford J, Garrigues N, et al. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria[J]. Applied and Environmental Microbiology, 2002, 68(4): 1786-1793. DOI:10.1128/AEM.68.4.1786-1793.2002 |
| [40] |
Aminov R, Garrigues-Jeanjean N, Mackie R. Molecular ecology of tetracycline resistance:Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins[J]. Applied and Environmental Microbiology, 2001, 67(1): 22-32. |
| [41] |
Zhang X X, Zhang T. Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across China and other global locations[J]. Environmental Science & Technology, 2011, 45(7): 2598-2604. |
| [42] |
Pei R T, Kim S C, Carlson K H, et al. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG)[J]. Water Research, 2006, 40(12): 2427-2435. DOI:10.1016/j.watres.2006.04.017 |
| [43] |
Pruden A, Pei R, Storteboom H, et al. Antibiotic resistance genes as emerging contaminants:Studies in Northern Colorado[J]. Environmental Science & Technology, 2006, 40(23): 7445-7450. |
| [44] |
Mu Q H, Li J, Sun Y X, et al. Occurrence of sulfonamide-, tetracycline-, plasmid-mediated quinolone-and macrolide-resistance genes in livestock feedlots in Northern China[J]. Environmental Science and Pollution Research, 2015, 22(9): 6932-6940. DOI:10.1007/s11356-014-3905-5 |
| [45] |
Kim H B, Wang M, Park C H, et al. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae[J]. Antimicrobial Agents and Chemotherapy, 2009, 53(8): 3582-3584. DOI:10.1128/AAC.01574-08 |
| [46] |
Xia L N, Li L, Wu C M, et al. A survey of plasmid-mediated fluoroquinolone resistance genes from Escherichia coli isolates and their dissemination in Shandong, China[J]. Foodborne Pathogens and Disease, 2010, 7(2): 207-215. DOI:10.1089/fpd.2009.0378 |
| [47] |
Cavaco L, Hasman H, Xia S, et al. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella entericaserovar Kentucky and Bovismorbificans strains of human origin[J]. Antimicrobial Agents and Chemotherapy, 2009, 53(2): 603-608. DOI:10.1128/AAC.00997-08 |
| [48] |
Cattoir V, Poirel L, Rotimi V, et al. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates[J]. Journal of Antimicrobial Chemotherapy, 2007, 60(2): 394-397. DOI:10.1093/jac/dkm204 |
| [49] |
Luo Y, Mao D Q, Rysz M, et al. Trends in antibiotic resistance genes occurrence in the Haihe River, China[J]. Environmental Science & Technology, 2010, 44(19): 7220-7225. |
| [50] |
Cheng W X, Chen H, Su C, et al. Abundance and persistence of antibiotic resistance genes in livestock farms:A comprehensive investigation in Eastern China[J]. Environmental International, 2013, 61: 1-7. DOI:10.1016/j.envint.2013.08.023 |
| [51] |
Zhao L, Dong Y H, Wang H. Residues of veterinary antibiotics in manures from feedlot livestock in eight provinces of China[J]. Science of the Total Environment, 2010, 408(5): 1069-1075. DOI:10.1016/j.scitotenv.2009.11.014 |
| [52] |
Roberts M C. Update on acquired tetracycline resistance genes[J]. FEMS Microbiology Letters, 2005, 245(2): 195-203. DOI:10.1016/j.femsle.2005.02.034 |
| [53] |
Akinbowale O L, Peng H, Barton M D. Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia[J]. Journal of Applied Microbiology, 2007, 103(5): 2016-2025. DOI:10.1111/j.1365-2672.2007.03445.x |
| [54] |
Rodriguez-Sanchez C, Altendorf K, Smalla K, et al. Spraying of oxytetracycline and gentamicin onto field-grown coriander did not affect the abundance of resistant bacteria, resistance genes, and broad host range plasmids detected in tropical soil bacteria[J]. Biology and Fertility of Soils, 2008, 44: 589-596. DOI:10.1007/s00374-007-0242-6 |
| [55] |
Roberts M C. Acquired tetracycline and/or macrolide-lincosamides-streptogramin resistance in anaerobes[J]. Anaerobe, 2003, 9(2): 63-69. DOI:10.1016/S1075-9964(03)00058-1 |
| [56] |
Liu Y F, Wang C H, Janapatla R P, et al. Presence of plasmid pa15 correlates with prevalence of constitutive mlsb resistance in group A Streptococcal isolates at a university hospital in Southern Taiwan[J]. Journal of Antimicrobial Chemotherapy, 2007, 59(2): 1167-1170. |
| [57] |
Luo Y, Wang Q, Lu Q, et al. An ionic liquid facilitates the proliferation of antibiotic resistance genes mediated by class Ⅰ integrons[J]. Environmental Science & Technology Letter, 2014, 1(5): 266-270. |
| [58] |
Hong B, Ba Y B, Niu L, et al. A comprehensive research on antibiotic resistance genes in microbiota of aquatic animals[J]. Frontiers in Microbiology, 2018, 9: 1617. DOI:10.3389/fmicb.2018.01617 |
| [59] |
Di Cesare A, Eckert E M, Rogora M, et al. Rainfall increases the abundance of antibiotic resistance genes within a riverine microbial community[J]. Environmental Pollution, 2017, 226: 473-478. DOI:10.1016/j.envpol.2017.04.036 |
| [60] |
Munir M, Xagoraraki I. Levels of antibiotic resistance genes in manure, biosolids, and fertilized soil[J]. Journal of Environmental Quality, 2011, 40(1): 248-255. |
| [61] |
Ji X L, Shen Q H, Liu F, et al. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai, China[J]. Journal of Hazardous Materials, 2012, 235(15): 178-185. |
| [62] |
Heuer H, Focks A, Lamshöft M, et al. Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil[J]. Soil Biology & Biochemistry, 2008, 40(7): 1892-1900. |
| [63] |
Heuer H, Kopmann C, Binh C T, et al. Spreading antibiotic resistance through spread manure:Characteristics of a novel plasmid type with low% G + C content[J]. Environmental Microbiology, 2009, 11(4): 937-949. DOI:10.1111/j.1462-2920.2008.01819.x |
| [64] |
Garder J L, Moorman T B, Soupir M L. Transport and persistence of tylosin-resistant enterococci, erm genes, and tylosin in soil and drainage water from fields receiving swine manure[J]. Journal of Environmental Quality, 2014, 43(4): 1484-1493. DOI:10.2134/jeq2013.09.0379 |
| [65] |
Michaud L, Lo Giudice A, Interdonato F, et al. C/N ratio-induced structural shift of bacterial communities inside lab-scale aquaculture biofilters[J]. Aquacultural Engineering, 2014, 58: 77-87. DOI:10.1016/j.aquaeng.2013.11.002 |
| [66] |
Awasthi M K, Liu T, Chen H, et al. The behavior of antibiotic resistance genes and their associations with bacterial community during poultry manure composting[J]. Bioresource Technology, 2019, 280: 70-78. DOI:10.1016/j.biortech.2019.02.030 |
| [67] |
Geisseler D, Horwath, W R. Relationship between carbon and nitrogen availability and extracellular enzyme activities in soil[J]. Pedobiologia, 2009, 53(1): 87-98. DOI:10.1016/j.pedobi.2009.06.002 |
| [68] |
Tets G V, Artemenko N K, Zaslavskaia N V, et al. Effect of exogenic proteolytic enzymes on transfer of plasmid genes in mixed bacterial biofilms[J]. Antibiotics and Chemoterapy, 2009, 3-5. |
| [69] |
Hu J, Li D P, Tao Y, et al. Microbial diversity of nitrite-oxidizing and heterotrophic bacterial communities under different C/N ratios[J]. Chinese Journal of Applied and Environmental Biology, 2009, 15(3): 351-355. |
| [70] |
任四伟.抗生素抗性细菌在粪便、土壤、蔬菜中的分布[D].新乡: 河南师范大学, 2013. REN Si-wei. Distribution of antibiotic resistant bacteria in feces, soil and fertilized vegetables[D]. Xinxiang: Henan Normal University, 2013. |
| [71] |
蒋志云, 韦佳敏, 缪新年, 等. ABR-MBR工艺反硝化除磷微生物群落特征分析[J]. 环境工程学报, 2019, 13(7): 1653-1661. JIANG Zhi-yun, WEI Jia-min, MIAO Xin-nian, et al. Analysis of microbial community characteristics of denitrifying phosphorus removal in the ABR-MBR process[J]. Chinese Journal of Environmental Engineering, 2019, 13(7): 1653-1661. |
| [72] |
Chironna M, Sallustio A, Esposito S, et al. Emergence of macrolideresistant strains during an outbreak of mycoplasma pneumoniae infections in children[J]. Journal of Antimicrobial Chemotherapy, 2011, 66: 734-737. DOI:10.1093/jac/dkr003 |
| [73] |
Nilsson A C, Jensen J S, Bjorkman P, et al. Development of macrolide resistance in mycoplasma pneumoniae-infected swedish patients treated with macrolides[J]. Scandinavian Journal of Infectious Diseases, 2014, 46: 315-319. DOI:10.3109/00365548.2013.866268 |
 2020, Vol. 39
2020, Vol. 39